| Full name: hyperpolarization activated cyclic nucleotide gated potassium channel 3 | Alias Symbol: KIAA1535 | ||
| Type: protein-coding gene | Cytoband: 1q22 | ||
| Entrez ID: 57657 | HGNC ID: HGNC:19183 | Ensembl Gene: ENSG00000143630 | OMIM ID: 609973 |
| Drug and gene relationship at DGIdb | |||
Expression of HCN3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCN3 | 57657 | 228741_s_at | 0.0847 | 0.7946 | |
| GSE26886 | HCN3 | 57657 | 228741_s_at | 0.0188 | 0.9068 | |
| GSE45670 | HCN3 | 57657 | 228741_s_at | 0.0591 | 0.6602 | |
| GSE63941 | HCN3 | 57657 | 228741_s_at | 0.2167 | 0.2475 | |
| GSE77861 | HCN3 | 57657 | 228741_s_at | 0.0247 | 0.8544 | |
| GSE97050 | HCN3 | 57657 | A_23_P34827 | 0.0466 | 0.8563 | |
| SRP007169 | HCN3 | 57657 | RNAseq | -0.9860 | 0.1430 | |
| SRP064894 | HCN3 | 57657 | RNAseq | 0.1085 | 0.6813 | |
| SRP133303 | HCN3 | 57657 | RNAseq | -0.1140 | 0.7190 | |
| SRP159526 | HCN3 | 57657 | RNAseq | 0.0620 | 0.9020 | |
| SRP193095 | HCN3 | 57657 | RNAseq | -0.0970 | 0.4975 | |
| SRP219564 | HCN3 | 57657 | RNAseq | 0.4488 | 0.5192 | |
| TCGA | HCN3 | 57657 | RNAseq | 0.2549 | 0.0055 |
Upregulated datasets: 0; Downregulated datasets: 0.
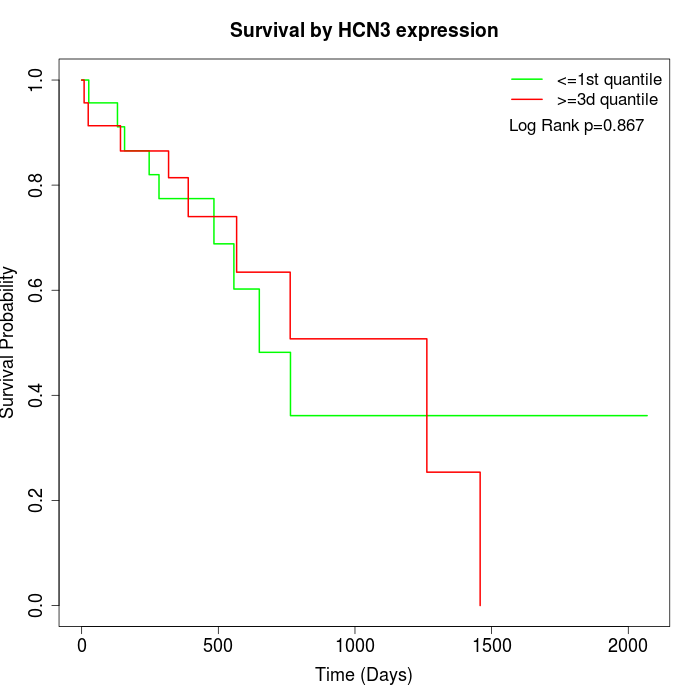
Survival by HCN3 expression:
Note: Click image to view full size file.
Copy number change of HCN3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCN3 | 57657 | 14 | 0 | 16 | |
| GSE20123 | HCN3 | 57657 | 13 | 0 | 17 | |
| GSE43470 | HCN3 | 57657 | 7 | 3 | 33 | |
| GSE46452 | HCN3 | 57657 | 2 | 1 | 56 | |
| GSE47630 | HCN3 | 57657 | 14 | 0 | 26 | |
| GSE54993 | HCN3 | 57657 | 0 | 4 | 66 | |
| GSE54994 | HCN3 | 57657 | 16 | 0 | 37 | |
| GSE60625 | HCN3 | 57657 | 0 | 0 | 11 | |
| GSE74703 | HCN3 | 57657 | 7 | 2 | 27 | |
| GSE74704 | HCN3 | 57657 | 6 | 0 | 14 | |
| TCGA | HCN3 | 57657 | 38 | 2 | 56 |
Total number of gains: 117; Total number of losses: 12; Total Number of normals: 359.
Somatic mutations of HCN3:
Generating mutation plots.
Highly correlated genes for HCN3:
Showing top 20/98 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCN3 | ARL8A | 0.801726 | 3 | 0 | 3 |
| HCN3 | KLRG2 | 0.729584 | 3 | 0 | 3 |
| HCN3 | PRKAR1B | 0.716304 | 3 | 0 | 3 |
| HCN3 | CNTD2 | 0.706329 | 3 | 0 | 3 |
| HCN3 | CPAMD8 | 0.696057 | 4 | 0 | 3 |
| HCN3 | SLC22A31 | 0.691807 | 3 | 0 | 3 |
| HCN3 | CNGB3 | 0.69073 | 3 | 0 | 3 |
| HCN3 | GSDMD | 0.679546 | 3 | 0 | 3 |
| HCN3 | TFAP2C | 0.66988 | 3 | 0 | 3 |
| HCN3 | C16orf95 | 0.66878 | 3 | 0 | 3 |
| HCN3 | SYCE1L | 0.663219 | 3 | 0 | 3 |
| HCN3 | HSF4 | 0.660878 | 3 | 0 | 3 |
| HCN3 | EPHA10 | 0.657941 | 3 | 0 | 3 |
| HCN3 | SYNGAP1 | 0.643588 | 4 | 0 | 3 |
| HCN3 | TBKBP1 | 0.642954 | 4 | 0 | 3 |
| HCN3 | PHF1 | 0.641867 | 3 | 0 | 3 |
| HCN3 | SPRED3 | 0.641639 | 5 | 0 | 3 |
| HCN3 | HDAC10 | 0.639532 | 3 | 0 | 3 |
| HCN3 | EYS | 0.638587 | 4 | 0 | 3 |
| HCN3 | LRFN4 | 0.638491 | 3 | 0 | 3 |
For details and further investigation, click here