| Full name: hes family bHLH transcription factor 1 | Alias Symbol: FLJ20408|HES-1|Hes1|bHLHb39 | ||
| Type: protein-coding gene | Cytoband: 3q29 | ||
| Entrez ID: 3280 | HGNC ID: HGNC:5192 | Ensembl Gene: ENSG00000114315 | OMIM ID: 139605 |
| Drug and gene relationship at DGIdb | |||
HES1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03460 | Fanconi anemia pathway | |
| hsa04330 | Notch signaling pathway | |
| hsa04950 | Maturity onset diabetes of the young |
Expression of HES1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HES1 | 3280 | 203394_s_at | -0.6576 | 0.3960 | |
| GSE20347 | HES1 | 3280 | 203394_s_at | -0.8998 | 0.0002 | |
| GSE23400 | HES1 | 3280 | 203394_s_at | -0.5628 | 0.0000 | |
| GSE26886 | HES1 | 3280 | 203394_s_at | -0.3562 | 0.3222 | |
| GSE29001 | HES1 | 3280 | 203394_s_at | -0.8762 | 0.0249 | |
| GSE38129 | HES1 | 3280 | 203394_s_at | -0.6074 | 0.0167 | |
| GSE45670 | HES1 | 3280 | 203394_s_at | 0.0255 | 0.9353 | |
| GSE53622 | HES1 | 3280 | 96542 | -0.4639 | 0.0000 | |
| GSE53624 | HES1 | 3280 | 96542 | -0.7593 | 0.0000 | |
| GSE63941 | HES1 | 3280 | 203394_s_at | 2.9855 | 0.0031 | |
| GSE77861 | HES1 | 3280 | 203394_s_at | -0.3915 | 0.3420 | |
| GSE97050 | HES1 | 3280 | A_23_P6596 | -0.2251 | 0.4804 | |
| SRP007169 | HES1 | 3280 | RNAseq | -1.8095 | 0.0001 | |
| SRP008496 | HES1 | 3280 | RNAseq | -1.7582 | 0.0000 | |
| SRP064894 | HES1 | 3280 | RNAseq | -0.5675 | 0.0435 | |
| SRP133303 | HES1 | 3280 | RNAseq | -0.1282 | 0.7175 | |
| SRP159526 | HES1 | 3280 | RNAseq | -0.7479 | 0.0380 | |
| SRP193095 | HES1 | 3280 | RNAseq | -0.5361 | 0.0004 | |
| SRP219564 | HES1 | 3280 | RNAseq | -0.9184 | 0.0644 | |
| TCGA | HES1 | 3280 | RNAseq | 0.2698 | 0.0002 |
Upregulated datasets: 1; Downregulated datasets: 2.
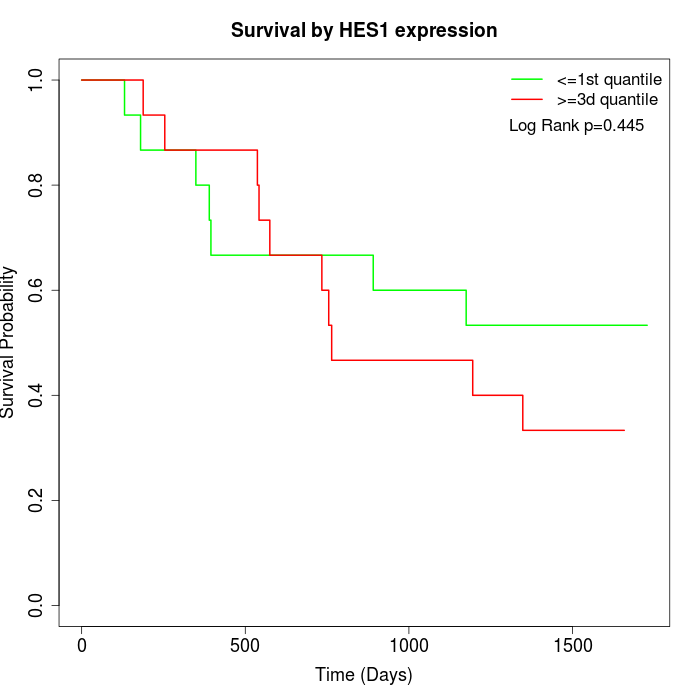
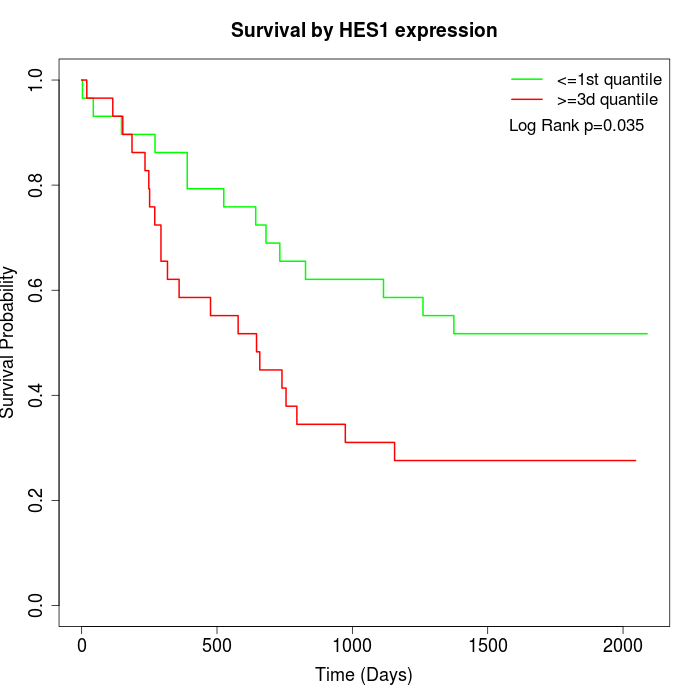
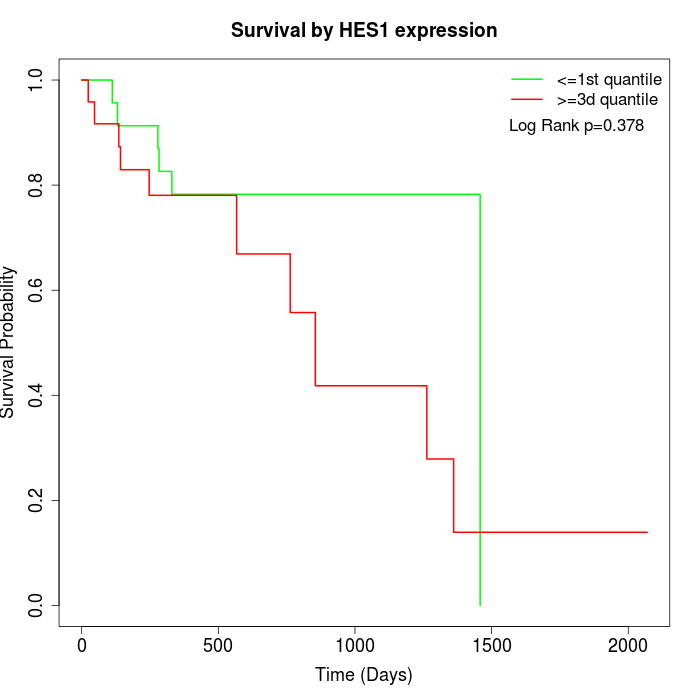
Survival by HES1 expression:
Note: Click image to view full size file.
Copy number change of HES1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HES1 | 3280 | 22 | 0 | 8 | |
| GSE20123 | HES1 | 3280 | 22 | 0 | 8 | |
| GSE43470 | HES1 | 3280 | 24 | 0 | 19 | |
| GSE46452 | HES1 | 3280 | 19 | 1 | 39 | |
| GSE47630 | HES1 | 3280 | 27 | 2 | 11 | |
| GSE54993 | HES1 | 3280 | 1 | 21 | 48 | |
| GSE54994 | HES1 | 3280 | 41 | 0 | 12 | |
| GSE60625 | HES1 | 3280 | 0 | 6 | 5 | |
| GSE74703 | HES1 | 3280 | 20 | 0 | 16 | |
| GSE74704 | HES1 | 3280 | 14 | 0 | 6 | |
| TCGA | HES1 | 3280 | 77 | 0 | 19 |
Total number of gains: 267; Total number of losses: 30; Total Number of normals: 191.
Somatic mutations of HES1:
Generating mutation plots.
Highly correlated genes for HES1:
Showing top 20/619 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HES1 | KAT7 | 0.727503 | 3 | 0 | 3 |
| HES1 | NRARP | 0.694153 | 4 | 0 | 4 |
| HES1 | SLC10A5 | 0.688138 | 3 | 0 | 3 |
| HES1 | RAB4B | 0.68471 | 3 | 0 | 3 |
| HES1 | KLRG1 | 0.682719 | 3 | 0 | 3 |
| HES1 | COPZ1 | 0.660961 | 3 | 0 | 3 |
| HES1 | GRPEL2 | 0.658345 | 3 | 0 | 3 |
| HES1 | SYN3 | 0.650758 | 3 | 0 | 3 |
| HES1 | EMP2 | 0.650701 | 7 | 0 | 6 |
| HES1 | IL7 | 0.650013 | 3 | 0 | 3 |
| HES1 | NAA20 | 0.649661 | 4 | 0 | 4 |
| HES1 | CA13 | 0.648428 | 3 | 0 | 3 |
| HES1 | PDCD4 | 0.647959 | 3 | 0 | 3 |
| HES1 | SCYL3 | 0.647583 | 3 | 0 | 3 |
| HES1 | CDC14A | 0.645433 | 3 | 0 | 3 |
| HES1 | CTNND1 | 0.644927 | 3 | 0 | 3 |
| HES1 | INTS6 | 0.644718 | 3 | 0 | 3 |
| HES1 | FAM110C | 0.640988 | 5 | 0 | 4 |
| HES1 | SPRYD4 | 0.640475 | 4 | 0 | 4 |
| HES1 | FAM135A | 0.634103 | 4 | 0 | 3 |
For details and further investigation, click here