| Full name: catenin delta 1 | Alias Symbol: KIAA0384|p120|p120cas|p120ctn | ||
| Type: protein-coding gene | Cytoband: 11q12.1 | ||
| Entrez ID: 1500 | HGNC ID: HGNC:2515 | Ensembl Gene: ENSG00000198561 | OMIM ID: 601045 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CTNND1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04520 | Adherens junction | |
| hsa04670 | Leukocyte transendothelial migration |
Expression of CTNND1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CTNND1 | 1500 | 128421 | -0.5951 | 0.0000 | |
| GSE53624 | CTNND1 | 1500 | 128421 | -0.6512 | 0.0000 | |
| GSE97050 | CTNND1 | 1500 | A_33_P3209716 | -0.0152 | 0.9760 | |
| SRP007169 | CTNND1 | 1500 | RNAseq | -0.2267 | 0.5351 | |
| SRP008496 | CTNND1 | 1500 | RNAseq | -0.5118 | 0.0263 | |
| SRP064894 | CTNND1 | 1500 | RNAseq | -1.2639 | 0.0000 | |
| SRP133303 | CTNND1 | 1500 | RNAseq | -0.5573 | 0.0080 | |
| SRP159526 | CTNND1 | 1500 | RNAseq | -0.8468 | 0.0005 | |
| SRP193095 | CTNND1 | 1500 | RNAseq | -0.9399 | 0.0000 | |
| SRP219564 | CTNND1 | 1500 | RNAseq | -1.1642 | 0.0404 | |
| TCGA | CTNND1 | 1500 | RNAseq | -0.0026 | 0.9554 |
Upregulated datasets: 0; Downregulated datasets: 2.
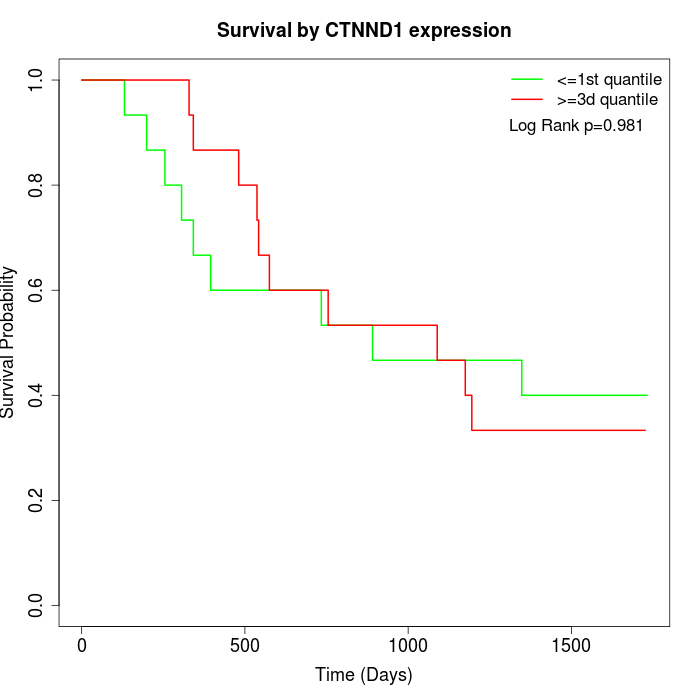
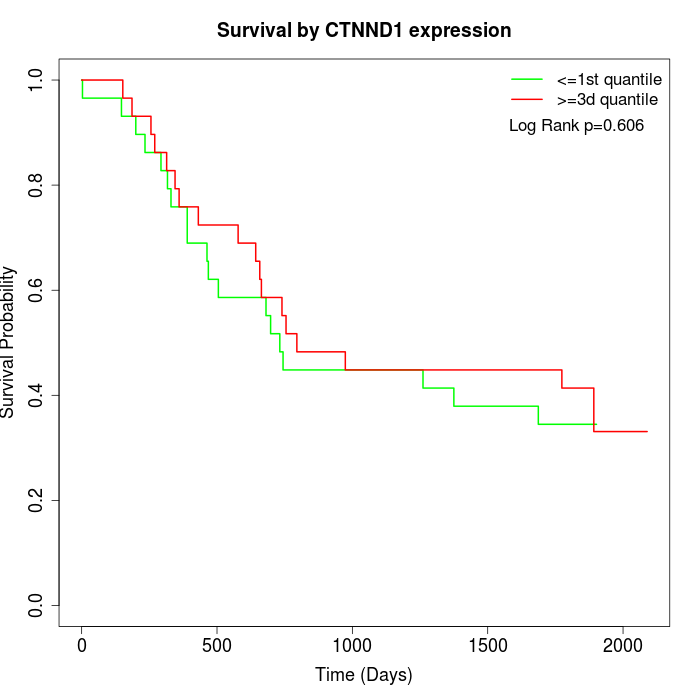
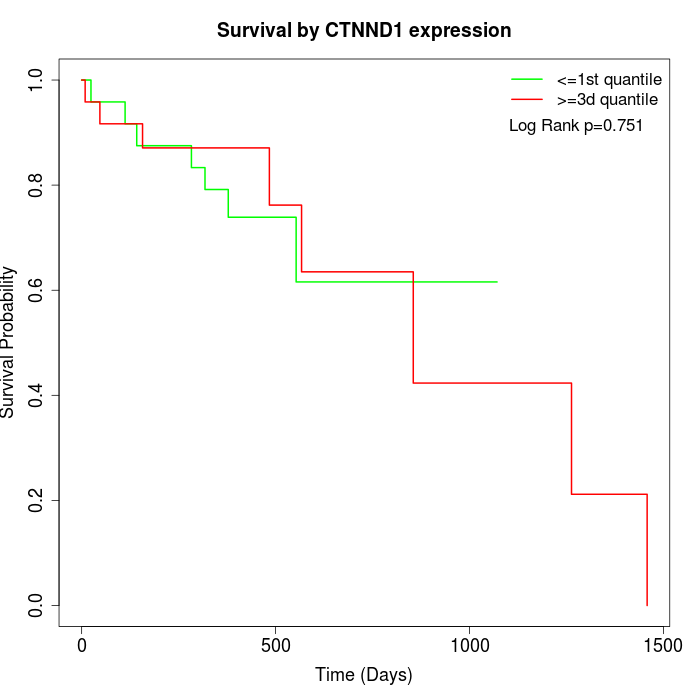
Survival by CTNND1 expression:
Note: Click image to view full size file.
Copy number change of CTNND1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTNND1 | 1500 | 2 | 3 | 25 | |
| GSE20123 | CTNND1 | 1500 | 2 | 3 | 25 | |
| GSE43470 | CTNND1 | 1500 | 1 | 5 | 37 | |
| GSE46452 | CTNND1 | 1500 | 9 | 4 | 46 | |
| GSE47630 | CTNND1 | 1500 | 3 | 8 | 29 | |
| GSE54993 | CTNND1 | 1500 | 3 | 0 | 67 | |
| GSE54994 | CTNND1 | 1500 | 5 | 6 | 42 | |
| GSE60625 | CTNND1 | 1500 | 0 | 3 | 8 | |
| GSE74703 | CTNND1 | 1500 | 1 | 3 | 32 | |
| GSE74704 | CTNND1 | 1500 | 2 | 3 | 15 | |
| TCGA | CTNND1 | 1500 | 14 | 10 | 72 |
Total number of gains: 42; Total number of losses: 48; Total Number of normals: 398.
Somatic mutations of CTNND1:
Generating mutation plots.
Highly correlated genes for CTNND1:
Showing top 20/323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTNND1 | ARHGAP32 | 0.829739 | 3 | 0 | 3 |
| CTNND1 | FAM135A | 0.803424 | 3 | 0 | 3 |
| CTNND1 | GRHL1 | 0.79218 | 3 | 0 | 3 |
| CTNND1 | MPZL2 | 0.791248 | 3 | 0 | 3 |
| CTNND1 | CAPN1 | 0.790572 | 3 | 0 | 3 |
| CTNND1 | S100A16 | 0.776598 | 3 | 0 | 3 |
| CTNND1 | ZDHHC13 | 0.76285 | 3 | 0 | 3 |
| CTNND1 | FAM160A1 | 0.762313 | 3 | 0 | 3 |
| CTNND1 | ARHGAP27 | 0.757153 | 3 | 0 | 3 |
| CTNND1 | DSG3 | 0.756219 | 3 | 0 | 3 |
| CTNND1 | TMEM40 | 0.755972 | 3 | 0 | 3 |
| CTNND1 | TGM1 | 0.755856 | 3 | 0 | 3 |
| CTNND1 | S100A14 | 0.755671 | 3 | 0 | 3 |
| CTNND1 | SERPINB1 | 0.751325 | 3 | 0 | 3 |
| CTNND1 | BTBD11 | 0.751007 | 3 | 0 | 3 |
| CTNND1 | NIPAL1 | 0.750687 | 3 | 0 | 3 |
| CTNND1 | EHF | 0.75052 | 3 | 0 | 3 |
| CTNND1 | RAB27B | 0.750442 | 3 | 0 | 3 |
| CTNND1 | ZNF750 | 0.748762 | 3 | 0 | 3 |
| CTNND1 | PGLYRP3 | 0.748002 | 3 | 0 | 3 |
For details and further investigation, click here