| Full name: hes family bHLH transcription factor 2 | Alias Symbol: bHLHb40 | ||
| Type: protein-coding gene | Cytoband: 1p36.31 | ||
| Entrez ID: 54626 | HGNC ID: HGNC:16005 | Ensembl Gene: ENSG00000069812 | OMIM ID: 609970 |
| Drug and gene relationship at DGIdb | |||
Expression of HES2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HES2 | 54626 | 231928_at | 0.4486 | 0.6622 | |
| GSE20347 | HES2 | 54626 | 214521_at | -0.2179 | 0.0106 | |
| GSE23400 | HES2 | 54626 | 214521_at | -0.0642 | 0.1261 | |
| GSE26886 | HES2 | 54626 | 231928_at | -0.0388 | 0.9360 | |
| GSE29001 | HES2 | 54626 | 216674_at | 0.0345 | 0.8174 | |
| GSE38129 | HES2 | 54626 | 216674_at | 0.0308 | 0.8186 | |
| GSE45670 | HES2 | 54626 | 231928_at | 0.6670 | 0.0177 | |
| GSE53622 | HES2 | 54626 | 116480 | 0.0999 | 0.7255 | |
| GSE53624 | HES2 | 54626 | 116480 | -0.1743 | 0.2346 | |
| GSE63941 | HES2 | 54626 | 231928_at | 1.0731 | 0.1345 | |
| GSE77861 | HES2 | 54626 | 231928_at | -0.1944 | 0.3671 | |
| GSE97050 | HES2 | 54626 | A_33_P3376365 | 0.2140 | 0.3437 | |
| SRP007169 | HES2 | 54626 | RNAseq | -0.6453 | 0.1658 | |
| SRP008496 | HES2 | 54626 | RNAseq | -0.9617 | 0.0034 | |
| SRP064894 | HES2 | 54626 | RNAseq | -0.4597 | 0.2756 | |
| SRP133303 | HES2 | 54626 | RNAseq | 0.1440 | 0.7189 | |
| SRP159526 | HES2 | 54626 | RNAseq | -1.0365 | 0.1535 | |
| SRP193095 | HES2 | 54626 | RNAseq | 0.0606 | 0.8685 | |
| SRP219564 | HES2 | 54626 | RNAseq | -0.1927 | 0.8617 | |
| TCGA | HES2 | 54626 | RNAseq | 1.7290 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
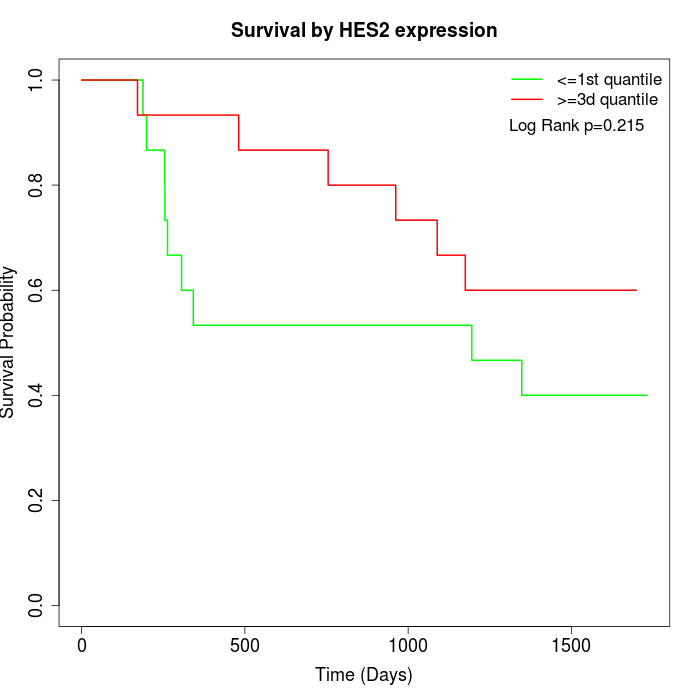
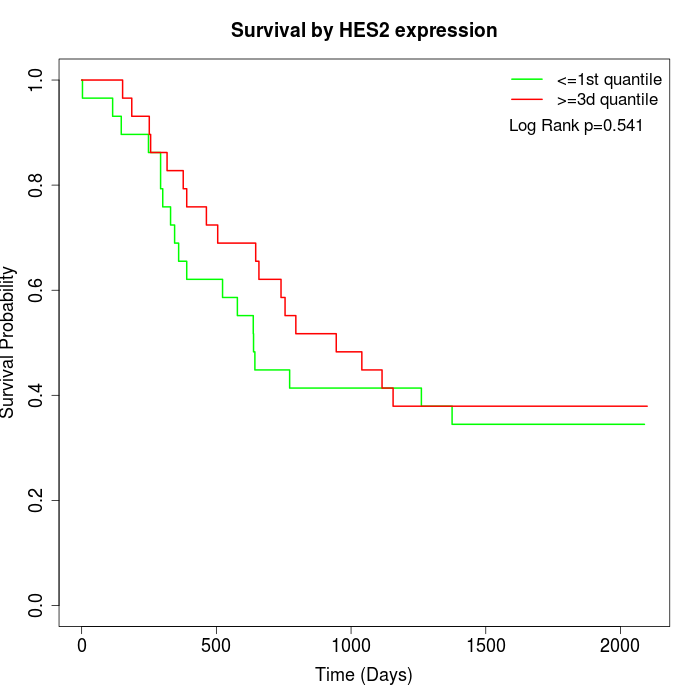
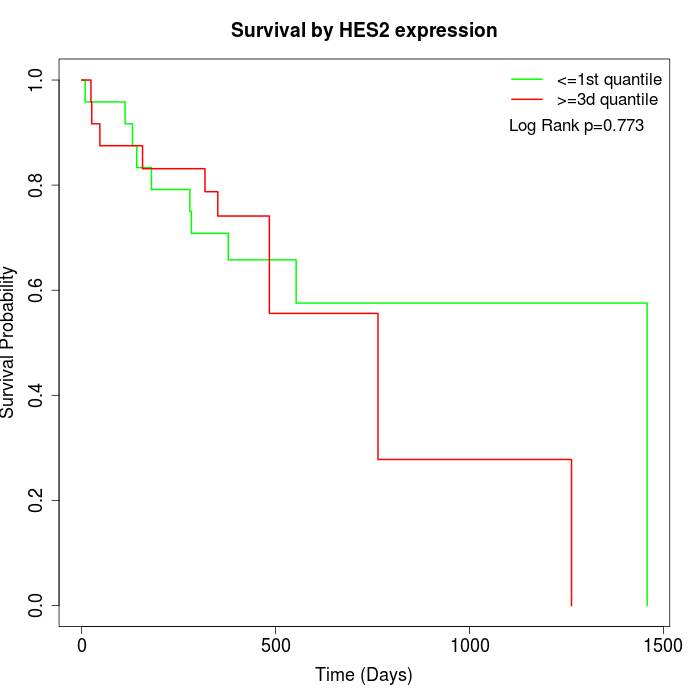
Survival by HES2 expression:
Note: Click image to view full size file.
Copy number change of HES2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HES2 | 54626 | 3 | 6 | 21 | |
| GSE20123 | HES2 | 54626 | 3 | 5 | 22 | |
| GSE43470 | HES2 | 54626 | 6 | 5 | 32 | |
| GSE46452 | HES2 | 54626 | 7 | 1 | 51 | |
| GSE47630 | HES2 | 54626 | 8 | 4 | 28 | |
| GSE54993 | HES2 | 54626 | 3 | 2 | 65 | |
| GSE54994 | HES2 | 54626 | 14 | 3 | 36 | |
| GSE60625 | HES2 | 54626 | 0 | 0 | 11 | |
| GSE74703 | HES2 | 54626 | 5 | 3 | 28 | |
| GSE74704 | HES2 | 54626 | 3 | 0 | 17 | |
| TCGA | HES2 | 54626 | 12 | 22 | 62 |
Total number of gains: 64; Total number of losses: 51; Total Number of normals: 373.
Somatic mutations of HES2:
Generating mutation plots.
Highly correlated genes for HES2:
Showing top 20/144 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HES2 | ALKBH2 | 0.739764 | 3 | 0 | 3 |
| HES2 | C8G | 0.730078 | 3 | 0 | 3 |
| HES2 | ZNRF2 | 0.716856 | 3 | 0 | 3 |
| HES2 | AGAP3 | 0.705026 | 3 | 0 | 3 |
| HES2 | MIXL1 | 0.682323 | 3 | 0 | 3 |
| HES2 | LIMD1 | 0.679515 | 3 | 0 | 3 |
| HES2 | TRIM41 | 0.673928 | 4 | 0 | 3 |
| HES2 | FAT2 | 0.670487 | 6 | 0 | 6 |
| HES2 | NRG4 | 0.658861 | 4 | 0 | 3 |
| HES2 | ABCC6 | 0.65524 | 4 | 0 | 4 |
| HES2 | PRPF6 | 0.652691 | 3 | 0 | 3 |
| HES2 | GCH1 | 0.641703 | 4 | 0 | 3 |
| HES2 | OTUD4 | 0.640531 | 3 | 0 | 3 |
| HES2 | CRYGC | 0.64022 | 3 | 0 | 3 |
| HES2 | TEX13A | 0.63586 | 4 | 0 | 3 |
| HES2 | COL27A1 | 0.635138 | 3 | 0 | 3 |
| HES2 | SURF2 | 0.633727 | 4 | 0 | 3 |
| HES2 | PTK2B | 0.630011 | 4 | 0 | 3 |
| HES2 | GNRH2 | 0.629242 | 4 | 0 | 3 |
| HES2 | SLC10A6 | 0.628462 | 3 | 0 | 3 |
For details and further investigation, click here