| Full name: heparan-alpha-glucosaminide N-acetyltransferase | Alias Symbol: FLJ32731|HGNAT | ||
| Type: protein-coding gene | Cytoband: 8p11.21-p11.1 | ||
| Entrez ID: 138050 | HGNC ID: HGNC:26527 | Ensembl Gene: ENSG00000165102 | OMIM ID: 610453 |
| Drug and gene relationship at DGIdb | |||
Expression of HGSNAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HGSNAT | 138050 | 218017_s_at | 0.0506 | 0.9040 | |
| GSE20347 | HGSNAT | 138050 | 218017_s_at | -0.2997 | 0.1912 | |
| GSE23400 | HGSNAT | 138050 | 218017_s_at | 0.1959 | 0.0151 | |
| GSE26886 | HGSNAT | 138050 | 218017_s_at | -0.5349 | 0.0275 | |
| GSE29001 | HGSNAT | 138050 | 218017_s_at | -0.0506 | 0.8994 | |
| GSE38129 | HGSNAT | 138050 | 218017_s_at | -0.2256 | 0.1580 | |
| GSE45670 | HGSNAT | 138050 | 218017_s_at | -0.3938 | 0.0077 | |
| GSE53622 | HGSNAT | 138050 | 115368 | -0.0238 | 0.8187 | |
| GSE53624 | HGSNAT | 138050 | 115368 | -0.2010 | 0.0030 | |
| GSE63941 | HGSNAT | 138050 | 218017_s_at | -1.8290 | 0.0020 | |
| GSE77861 | HGSNAT | 138050 | 218017_s_at | -0.0204 | 0.9620 | |
| GSE97050 | HGSNAT | 138050 | A_23_P112061 | -0.1965 | 0.5359 | |
| SRP007169 | HGSNAT | 138050 | RNAseq | -0.0921 | 0.7780 | |
| SRP008496 | HGSNAT | 138050 | RNAseq | 0.2743 | 0.2792 | |
| SRP064894 | HGSNAT | 138050 | RNAseq | 0.2593 | 0.1884 | |
| SRP133303 | HGSNAT | 138050 | RNAseq | 0.1276 | 0.4059 | |
| SRP159526 | HGSNAT | 138050 | RNAseq | 0.1783 | 0.6841 | |
| SRP193095 | HGSNAT | 138050 | RNAseq | 0.1266 | 0.1684 | |
| SRP219564 | HGSNAT | 138050 | RNAseq | 0.0453 | 0.8503 | |
| TCGA | HGSNAT | 138050 | RNAseq | -0.0760 | 0.1001 |
Upregulated datasets: 0; Downregulated datasets: 1.
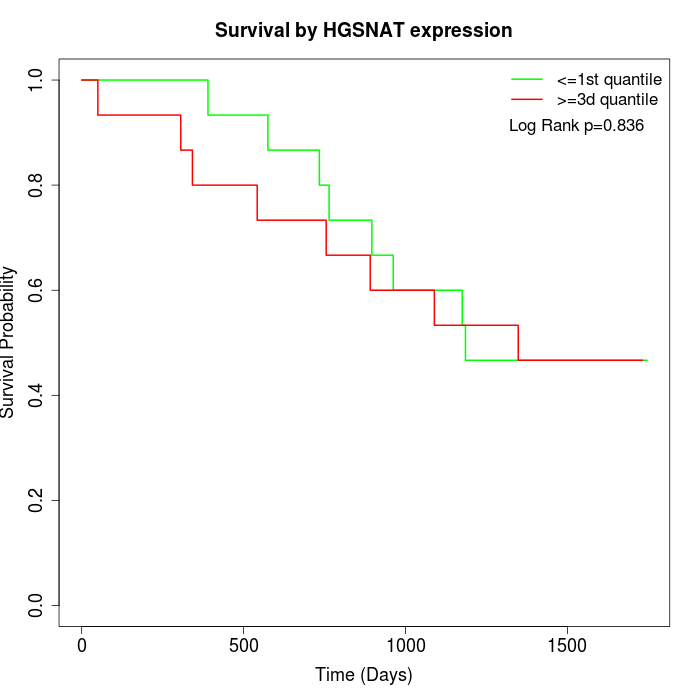
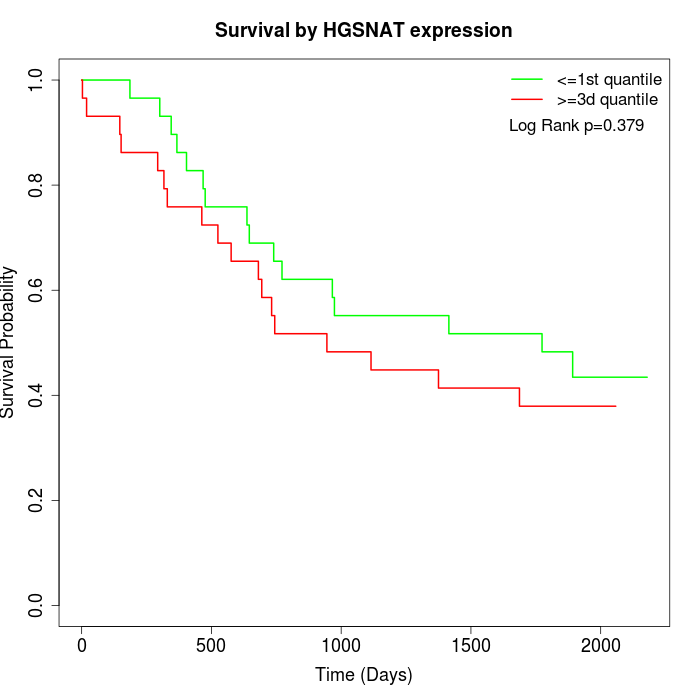
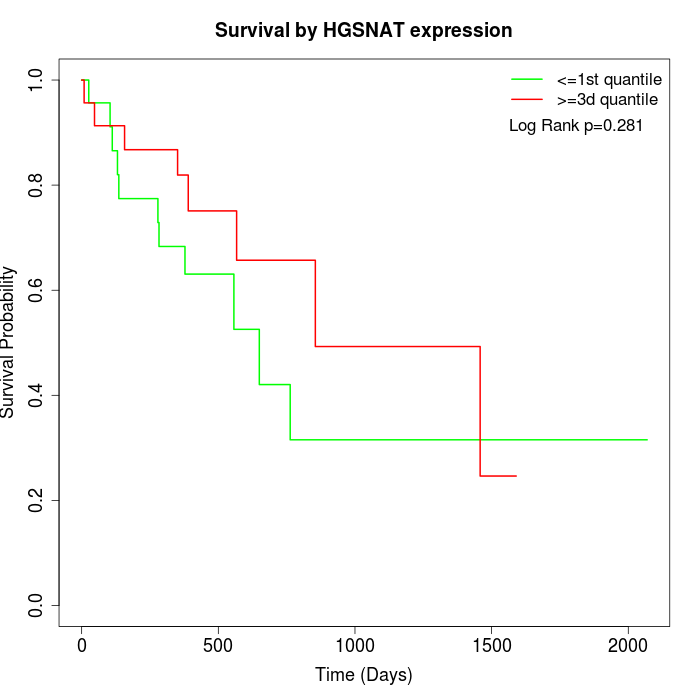
Survival by HGSNAT expression:
Note: Click image to view full size file.
Copy number change of HGSNAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HGSNAT | 138050 | 10 | 4 | 16 | |
| GSE20123 | HGSNAT | 138050 | 10 | 4 | 16 | |
| GSE43470 | HGSNAT | 138050 | 8 | 6 | 29 | |
| GSE46452 | HGSNAT | 138050 | 20 | 5 | 34 | |
| GSE47630 | HGSNAT | 138050 | 22 | 1 | 17 | |
| GSE54993 | HGSNAT | 138050 | 1 | 16 | 53 | |
| GSE54994 | HGSNAT | 138050 | 18 | 8 | 27 | |
| GSE60625 | HGSNAT | 138050 | 3 | 0 | 8 | |
| GSE74703 | HGSNAT | 138050 | 7 | 5 | 24 | |
| GSE74704 | HGSNAT | 138050 | 9 | 1 | 10 | |
| TCGA | HGSNAT | 138050 | 47 | 15 | 34 |
Total number of gains: 155; Total number of losses: 65; Total Number of normals: 268.
Somatic mutations of HGSNAT:
Generating mutation plots.
Highly correlated genes for HGSNAT:
Showing top 20/276 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HGSNAT | REEP3 | 0.73264 | 3 | 0 | 3 |
| HGSNAT | SIKE1 | 0.72598 | 4 | 0 | 4 |
| HGSNAT | RAP1A | 0.724035 | 3 | 0 | 3 |
| HGSNAT | BBS9 | 0.720652 | 3 | 0 | 3 |
| HGSNAT | NR2C2 | 0.714803 | 3 | 0 | 3 |
| HGSNAT | RRAGA | 0.697919 | 3 | 0 | 3 |
| HGSNAT | TK2 | 0.694784 | 4 | 0 | 4 |
| HGSNAT | DSTN | 0.69008 | 4 | 0 | 3 |
| HGSNAT | HDAC11 | 0.686346 | 3 | 0 | 3 |
| HGSNAT | PRKCA | 0.681056 | 3 | 0 | 3 |
| HGSNAT | CXCL12 | 0.677749 | 4 | 0 | 4 |
| HGSNAT | CUL2 | 0.677124 | 3 | 0 | 3 |
| HGSNAT | SNX18 | 0.676653 | 4 | 0 | 3 |
| HGSNAT | SORBS3 | 0.675224 | 3 | 0 | 3 |
| HGSNAT | CASP3 | 0.672898 | 3 | 0 | 3 |
| HGSNAT | ANXA11 | 0.669434 | 3 | 0 | 3 |
| HGSNAT | PRMT6 | 0.668086 | 3 | 0 | 3 |
| HGSNAT | NTN4 | 0.66719 | 3 | 0 | 3 |
| HGSNAT | EHBP1 | 0.665851 | 4 | 0 | 4 |
| HGSNAT | TBC1D14 | 0.664975 | 3 | 0 | 3 |
For details and further investigation, click here