| Full name: HHIP antisense RNA 1 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 4q31.21 | ||
| Entrez ID: 646576 | HGNC ID: HGNC:44182 | Ensembl Gene: ENSG00000248890 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of HHIP-AS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HHIP-AS1 | 646576 | 236632_at | -0.0539 | 0.8905 | |
| GSE26886 | HHIP-AS1 | 646576 | 236632_at | -0.1576 | 0.2131 | |
| GSE45670 | HHIP-AS1 | 646576 | 236632_at | -0.1344 | 0.1927 | |
| GSE53622 | HHIP-AS1 | 646576 | 43268 | 0.7941 | 0.0001 | |
| GSE53624 | HHIP-AS1 | 646576 | 43268 | 0.7042 | 0.0000 | |
| GSE63941 | HHIP-AS1 | 646576 | 236632_at | 0.2085 | 0.2686 | |
| GSE77861 | HHIP-AS1 | 646576 | 236632_at | -0.2489 | 0.1223 |
Upregulated datasets: 0; Downregulated datasets: 0.
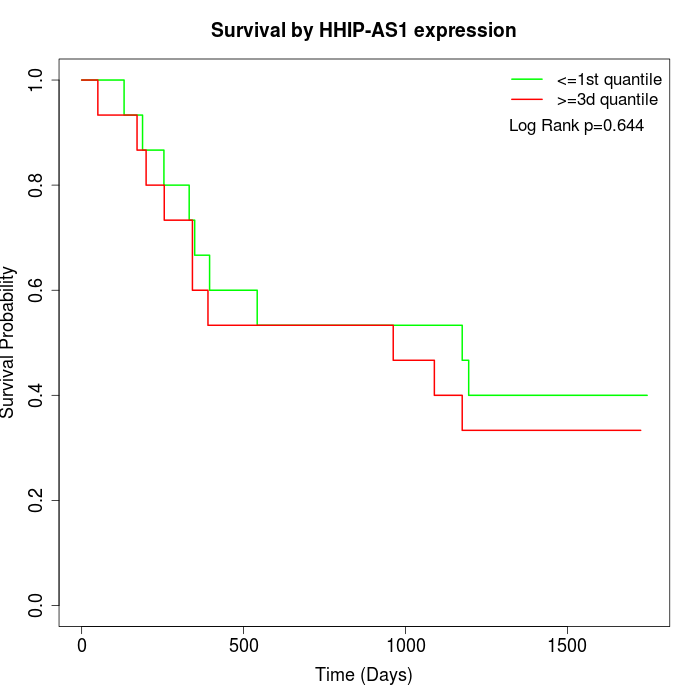
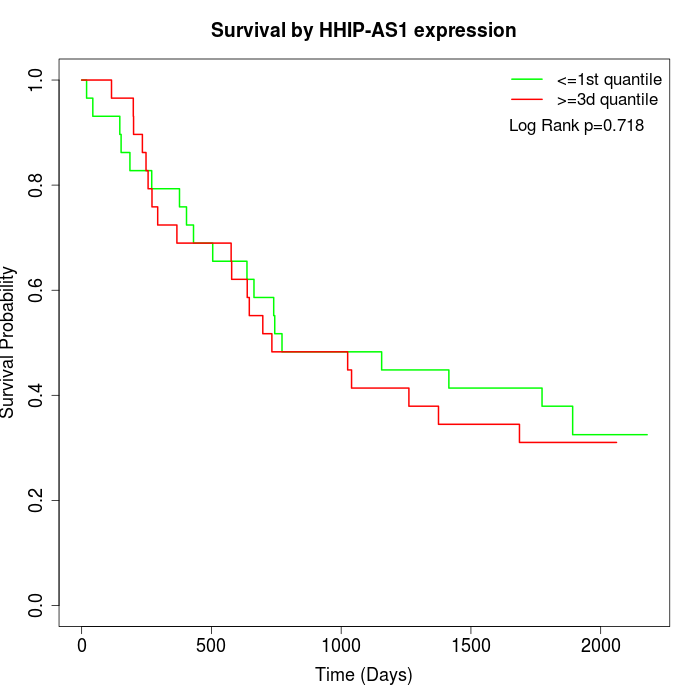
Survival by HHIP-AS1 expression:
Note: Click image to view full size file.
Copy number change of HHIP-AS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HHIP-AS1 | 646576 | 0 | 12 | 18 | |
| GSE20123 | HHIP-AS1 | 646576 | 0 | 12 | 18 | |
| GSE43470 | HHIP-AS1 | 646576 | 0 | 13 | 30 | |
| GSE46452 | HHIP-AS1 | 646576 | 1 | 36 | 22 | |
| GSE47630 | HHIP-AS1 | 646576 | 0 | 22 | 18 | |
| GSE54993 | HHIP-AS1 | 646576 | 10 | 0 | 60 | |
| GSE54994 | HHIP-AS1 | 646576 | 2 | 11 | 40 | |
| GSE60625 | HHIP-AS1 | 646576 | 0 | 1 | 10 | |
| GSE74703 | HHIP-AS1 | 646576 | 0 | 11 | 25 | |
| GSE74704 | HHIP-AS1 | 646576 | 0 | 6 | 14 |
Total number of gains: 13; Total number of losses: 124; Total Number of normals: 255.
Somatic mutations of HHIP-AS1:
Generating mutation plots.
Highly correlated genes for HHIP-AS1:
Showing all 19 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HHIP-AS1 | DPYSL4 | 0.64231 | 3 | 0 | 3 |
| HHIP-AS1 | GPR26 | 0.639479 | 3 | 0 | 3 |
| HHIP-AS1 | PTGER3 | 0.630337 | 4 | 0 | 3 |
| HHIP-AS1 | WDR86 | 0.627124 | 4 | 0 | 4 |
| HHIP-AS1 | ABCC8 | 0.624772 | 3 | 0 | 3 |
| HHIP-AS1 | PROCA1 | 0.620147 | 4 | 0 | 3 |
| HHIP-AS1 | HHIP | 0.615151 | 6 | 0 | 4 |
| HHIP-AS1 | PRKACG | 0.606504 | 3 | 0 | 3 |
| HHIP-AS1 | CDK14 | 0.589959 | 3 | 0 | 3 |
| HHIP-AS1 | IGSF22 | 0.583412 | 3 | 0 | 3 |
| HHIP-AS1 | BANF2 | 0.571153 | 4 | 0 | 3 |
| HHIP-AS1 | HTR1E | 0.565512 | 3 | 0 | 3 |
| HHIP-AS1 | CISH | 0.564701 | 3 | 0 | 3 |
| HHIP-AS1 | GLI1 | 0.560476 | 3 | 0 | 3 |
| HHIP-AS1 | FAM47C | 0.556814 | 4 | 0 | 3 |
| HHIP-AS1 | LINC00906 | 0.543281 | 3 | 0 | 3 |
| HHIP-AS1 | CNGB1 | 0.513518 | 4 | 0 | 3 |
| HHIP-AS1 | TMEM204 | 0.511324 | 6 | 0 | 4 |
| HHIP-AS1 | LGI4 | 0.502214 | 4 | 0 | 3 |
For details and further investigation, click here