| Full name: major histocompatibility complex, class I, E | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p22.1 | ||
| Entrez ID: 3133 | HGNC ID: HGNC:4962 | Ensembl Gene: ENSG00000204592 | OMIM ID: 143010 |
| Drug and gene relationship at DGIdb | |||
HLA-E involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa05166 | HTLV-I infection | |
| hsa05169 | Epstein-Barr virus infection |
Expression of HLA-E:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HLA-E | 3133 | 200905_x_at | 0.0072 | 0.9941 | |
| GSE20347 | HLA-E | 3133 | 200905_x_at | -0.3828 | 0.0355 | |
| GSE23400 | HLA-E | 3133 | 200905_x_at | -0.0273 | 0.8043 | |
| GSE26886 | HLA-E | 3133 | 200905_x_at | -0.7886 | 0.0041 | |
| GSE29001 | HLA-E | 3133 | 200905_x_at | -0.0197 | 0.9564 | |
| GSE38129 | HLA-E | 3133 | 200905_x_at | -0.2734 | 0.1325 | |
| GSE45670 | HLA-E | 3133 | 200905_x_at | 0.1969 | 0.3585 | |
| GSE53622 | HLA-E | 3133 | 87220 | 0.3172 | 0.0000 | |
| GSE53624 | HLA-E | 3133 | 87220 | 0.4160 | 0.0000 | |
| GSE63941 | HLA-E | 3133 | 200905_x_at | 0.0857 | 0.8875 | |
| GSE77861 | HLA-E | 3133 | 200905_x_at | -0.2386 | 0.4711 | |
| GSE97050 | HLA-E | 3133 | A_32_P460973 | 0.3235 | 0.2371 | |
| SRP007169 | HLA-E | 3133 | RNAseq | -0.9252 | 0.0180 | |
| SRP008496 | HLA-E | 3133 | RNAseq | -0.7284 | 0.0055 | |
| SRP064894 | HLA-E | 3133 | RNAseq | 0.0959 | 0.5949 | |
| SRP133303 | HLA-E | 3133 | RNAseq | -0.1679 | 0.3865 | |
| SRP159526 | HLA-E | 3133 | RNAseq | -0.6813 | 0.1902 | |
| SRP193095 | HLA-E | 3133 | RNAseq | -0.1758 | 0.2241 | |
| SRP219564 | HLA-E | 3133 | RNAseq | 0.1513 | 0.6563 | |
| TCGA | HLA-E | 3133 | RNAseq | 0.0661 | 0.2418 |
Upregulated datasets: 0; Downregulated datasets: 0.
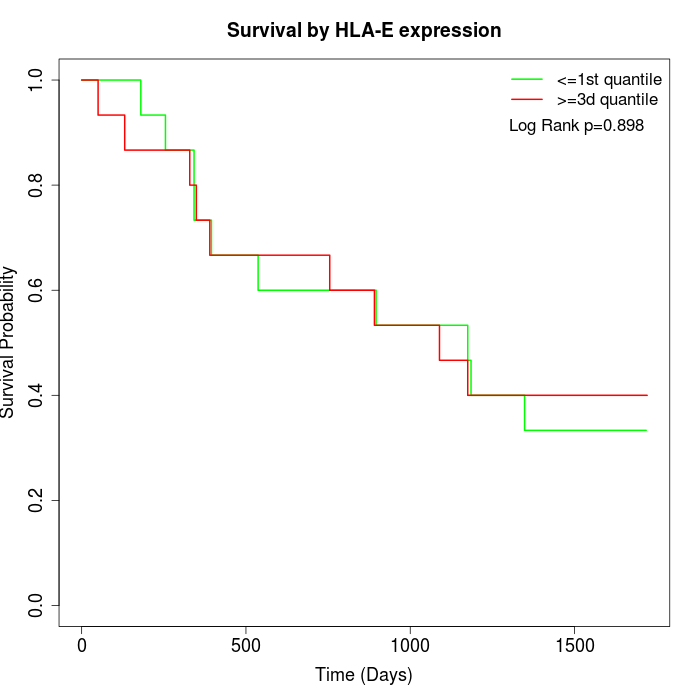
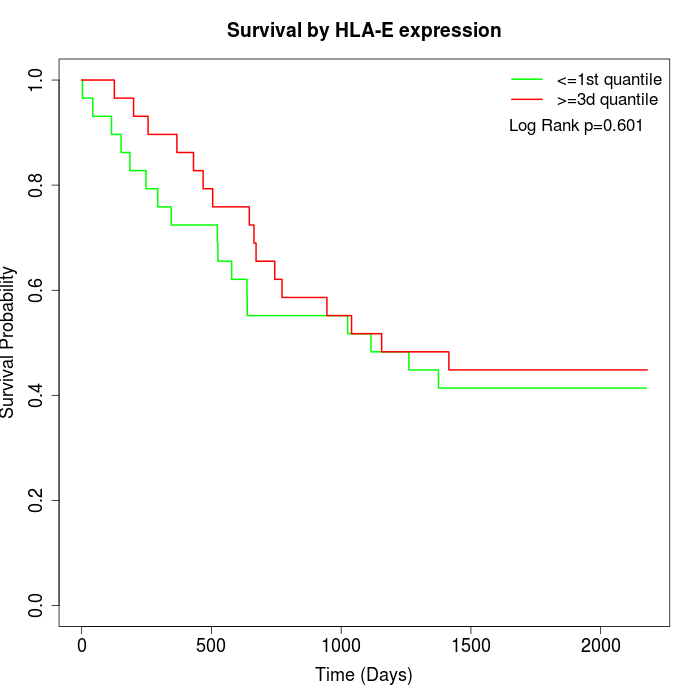
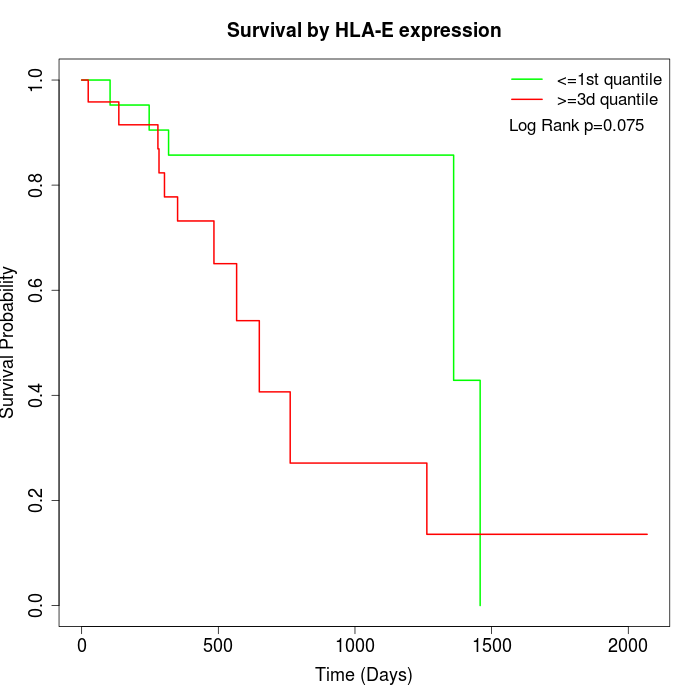
Survival by HLA-E expression:
Note: Click image to view full size file.
Copy number change of HLA-E:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HLA-E | 3133 | 5 | 1 | 24 | |
| GSE20123 | HLA-E | 3133 | 5 | 1 | 24 | |
| GSE43470 | HLA-E | 3133 | 5 | 1 | 37 | |
| GSE46452 | HLA-E | 3133 | 1 | 10 | 48 | |
| GSE47630 | HLA-E | 3133 | 7 | 3 | 30 | |
| GSE54993 | HLA-E | 3133 | 2 | 1 | 67 | |
| GSE54994 | HLA-E | 3133 | 11 | 4 | 38 | |
| GSE60625 | HLA-E | 3133 | 0 | 1 | 10 | |
| GSE74703 | HLA-E | 3133 | 5 | 0 | 31 | |
| GSE74704 | HLA-E | 3133 | 2 | 0 | 18 |
Total number of gains: 43; Total number of losses: 22; Total Number of normals: 327.
Somatic mutations of HLA-E:
Generating mutation plots.
Highly correlated genes for HLA-E:
Showing top 20/307 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HLA-E | PGK1 | 0.782981 | 3 | 0 | 3 |
| HLA-E | VCP | 0.751686 | 3 | 0 | 3 |
| HLA-E | PNPLA6 | 0.744645 | 3 | 0 | 3 |
| HLA-E | TPD52L2 | 0.728756 | 3 | 0 | 3 |
| HLA-E | WDR37 | 0.727132 | 3 | 0 | 3 |
| HLA-E | MAN2B2 | 0.720826 | 4 | 0 | 4 |
| HLA-E | DOK1 | 0.719303 | 3 | 0 | 3 |
| HLA-E | RPS19 | 0.719162 | 3 | 0 | 3 |
| HLA-E | HDAC3 | 0.715938 | 3 | 0 | 3 |
| HLA-E | GPR132 | 0.714086 | 3 | 0 | 3 |
| HLA-E | NDUFS2 | 0.710079 | 3 | 0 | 3 |
| HLA-E | DDX60L | 0.703111 | 4 | 0 | 4 |
| HLA-E | TRAF6 | 0.701938 | 3 | 0 | 3 |
| HLA-E | RAB2A | 0.69468 | 3 | 0 | 3 |
| HLA-E | SLC25A1 | 0.691195 | 3 | 0 | 3 |
| HLA-E | EPSTI1 | 0.689367 | 6 | 0 | 5 |
| HLA-E | STK38 | 0.686723 | 3 | 0 | 3 |
| HLA-E | HM13 | 0.685879 | 3 | 0 | 3 |
| HLA-E | TPRG1L | 0.685035 | 3 | 0 | 3 |
| HLA-E | ABLIM2 | 0.67653 | 3 | 0 | 3 |
For details and further investigation, click here