| Full name: phosphoglycerate kinase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xq21.1 | ||
| Entrez ID: 5230 | HGNC ID: HGNC:8896 | Ensembl Gene: ENSG00000102144 | OMIM ID: 311800 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PGK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of PGK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGK1 | 5230 | 200738_s_at | -0.1317 | 0.8213 | |
| GSE20347 | PGK1 | 5230 | 200738_s_at | -0.0317 | 0.9027 | |
| GSE23400 | PGK1 | 5230 | 200738_s_at | 0.1551 | 0.0388 | |
| GSE26886 | PGK1 | 5230 | 200738_s_at | -0.3896 | 0.0095 | |
| GSE29001 | PGK1 | 5230 | 200738_s_at | -0.3648 | 0.3592 | |
| GSE38129 | PGK1 | 5230 | 200738_s_at | 0.2204 | 0.1877 | |
| GSE45670 | PGK1 | 5230 | 200738_s_at | 0.1856 | 0.2679 | |
| GSE53622 | PGK1 | 5230 | 52666 | 0.1960 | 0.0510 | |
| GSE53624 | PGK1 | 5230 | 52666 | 0.1699 | 0.0207 | |
| GSE63941 | PGK1 | 5230 | 200738_s_at | 0.2064 | 0.6358 | |
| GSE77861 | PGK1 | 5230 | 200738_s_at | 0.0860 | 0.7824 | |
| GSE97050 | PGK1 | 5230 | A_23_P125829 | 0.1900 | 0.4258 | |
| SRP007169 | PGK1 | 5230 | RNAseq | 0.7502 | 0.2383 | |
| SRP008496 | PGK1 | 5230 | RNAseq | 0.7171 | 0.0630 | |
| SRP064894 | PGK1 | 5230 | RNAseq | 0.0286 | 0.8983 | |
| SRP133303 | PGK1 | 5230 | RNAseq | 0.6241 | 0.0220 | |
| SRP159526 | PGK1 | 5230 | RNAseq | -0.1590 | 0.6935 | |
| SRP193095 | PGK1 | 5230 | RNAseq | -0.0484 | 0.8029 | |
| SRP219564 | PGK1 | 5230 | RNAseq | -0.2766 | 0.4039 | |
| TCGA | PGK1 | 5230 | RNAseq | 0.2020 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
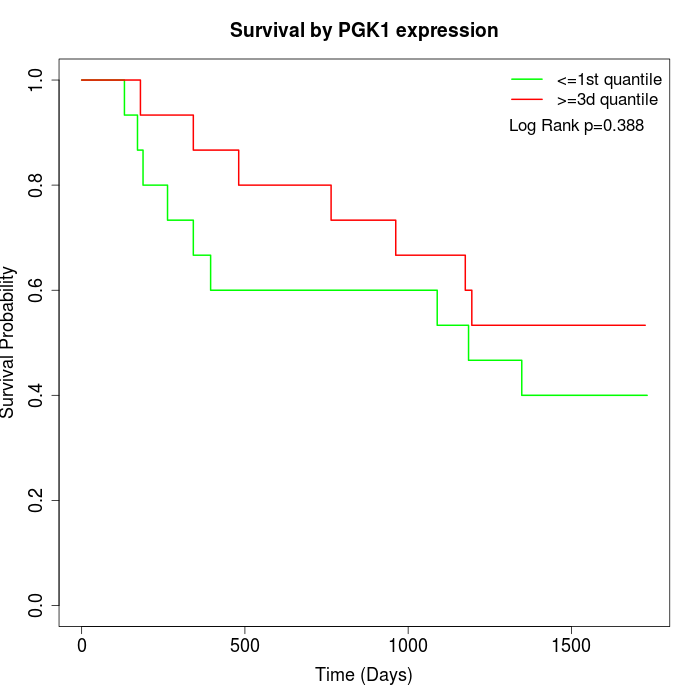
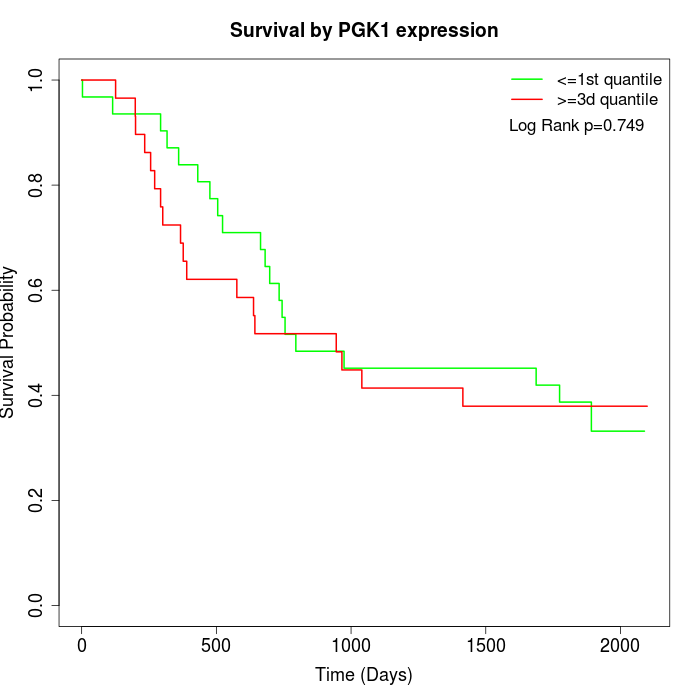
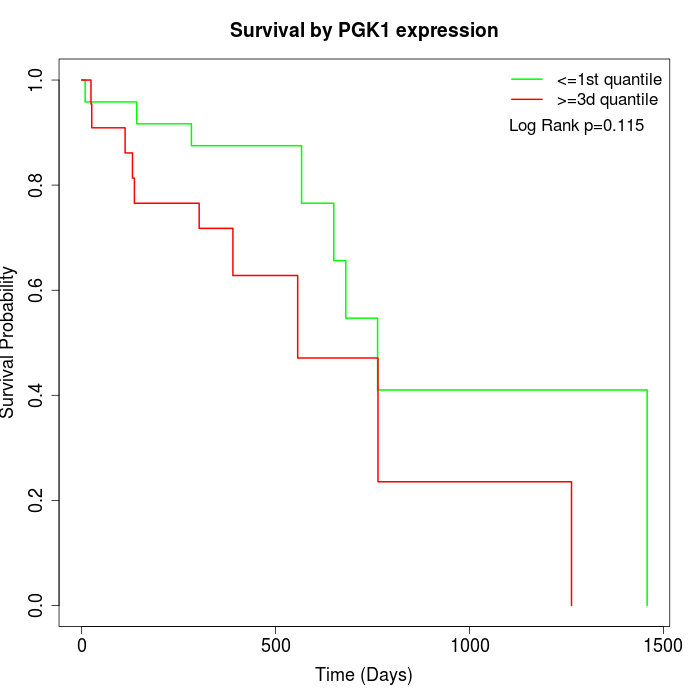
Survival by PGK1 expression:
Note: Click image to view full size file.
Copy number change of PGK1:
No record found for this gene.
Somatic mutations of PGK1:
Generating mutation plots.
Highly correlated genes for PGK1:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGK1 | HLA-E | 0.782981 | 3 | 0 | 3 |
| PGK1 | CMTM6 | 0.779287 | 3 | 0 | 3 |
| PGK1 | LAPTM4A | 0.764673 | 3 | 0 | 3 |
| PGK1 | DCTN3 | 0.736381 | 3 | 0 | 3 |
| PGK1 | SLC30A9 | 0.736345 | 3 | 0 | 3 |
| PGK1 | GPCPD1 | 0.715043 | 3 | 0 | 3 |
| PGK1 | SLAMF7 | 0.710424 | 3 | 0 | 3 |
| PGK1 | ATG12 | 0.708433 | 3 | 0 | 3 |
| PGK1 | GSDMC | 0.699631 | 4 | 0 | 4 |
| PGK1 | LACTB | 0.698002 | 3 | 0 | 3 |
| PGK1 | MAPK1IP1L | 0.696873 | 3 | 0 | 3 |
| PGK1 | FBXO3 | 0.694196 | 4 | 0 | 3 |
| PGK1 | GBE1 | 0.693617 | 4 | 0 | 3 |
| PGK1 | ATP6V1G1 | 0.685612 | 3 | 0 | 3 |
| PGK1 | PGM1 | 0.685205 | 3 | 0 | 3 |
| PGK1 | NLRP3 | 0.681441 | 3 | 0 | 3 |
| PGK1 | UGP2 | 0.677566 | 4 | 0 | 3 |
| PGK1 | WDR11 | 0.674342 | 3 | 0 | 3 |
| PGK1 | SCP2 | 0.671621 | 3 | 0 | 3 |
| PGK1 | TMED10 | 0.67142 | 4 | 0 | 4 |
For details and further investigation, click here