| Full name: homeobox C9 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.13 | ||
| Entrez ID: 3225 | HGNC ID: HGNC:5130 | Ensembl Gene: ENSG00000180806 | OMIM ID: 142971 |
| Drug and gene relationship at DGIdb | |||
Expression of HOXC9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HOXC9 | 3225 | 231936_at | 1.2443 | 0.0152 | |
| GSE26886 | HOXC9 | 3225 | 231936_at | 1.5935 | 0.0000 | |
| GSE45670 | HOXC9 | 3225 | 231936_at | 1.4148 | 0.0000 | |
| GSE63941 | HOXC9 | 3225 | 231936_at | 1.5181 | 0.0048 | |
| GSE77861 | HOXC9 | 3225 | 231936_at | 0.5791 | 0.0011 | |
| GSE97050 | HOXC9 | 3225 | A_23_P25150 | 0.4072 | 0.2017 | |
| SRP133303 | HOXC9 | 3225 | RNAseq | 3.0479 | 0.0000 | |
| SRP159526 | HOXC9 | 3225 | RNAseq | 3.6589 | 0.0000 | |
| SRP219564 | HOXC9 | 3225 | RNAseq | 2.9977 | 0.0001 | |
| TCGA | HOXC9 | 3225 | RNAseq | 3.0071 | 0.0000 |
Upregulated datasets: 8; Downregulated datasets: 0.
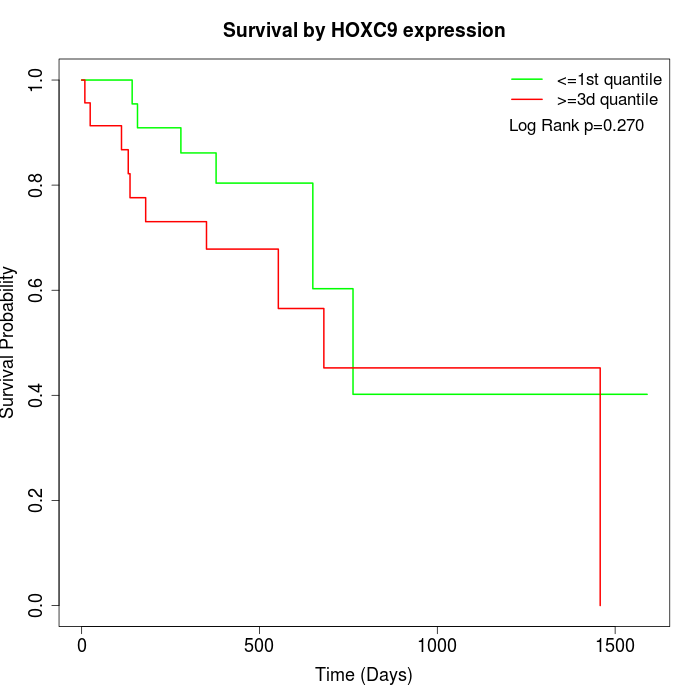
Survival by HOXC9 expression:
Note: Click image to view full size file.
Copy number change of HOXC9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HOXC9 | 3225 | 4 | 1 | 25 | |
| GSE20123 | HOXC9 | 3225 | 4 | 1 | 25 | |
| GSE43470 | HOXC9 | 3225 | 3 | 0 | 40 | |
| GSE46452 | HOXC9 | 3225 | 7 | 1 | 51 | |
| GSE47630 | HOXC9 | 3225 | 10 | 2 | 28 | |
| GSE54993 | HOXC9 | 3225 | 0 | 5 | 65 | |
| GSE54994 | HOXC9 | 3225 | 4 | 1 | 48 | |
| GSE60625 | HOXC9 | 3225 | 0 | 0 | 11 | |
| GSE74703 | HOXC9 | 3225 | 3 | 0 | 33 | |
| GSE74704 | HOXC9 | 3225 | 3 | 1 | 16 | |
| TCGA | HOXC9 | 3225 | 15 | 11 | 70 |
Total number of gains: 53; Total number of losses: 23; Total Number of normals: 412.
Somatic mutations of HOXC9:
Generating mutation plots.
Highly correlated genes for HOXC9:
Showing top 20/569 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HOXC9 | SDK1 | 0.821978 | 3 | 0 | 3 |
| HOXC9 | LONP1 | 0.819878 | 5 | 0 | 5 |
| HOXC9 | SEC62 | 0.815487 | 3 | 0 | 3 |
| HOXC9 | ARHGAP22 | 0.809306 | 3 | 0 | 3 |
| HOXC9 | CHST10 | 0.799517 | 3 | 0 | 3 |
| HOXC9 | SMG7 | 0.795411 | 4 | 0 | 4 |
| HOXC9 | MED20 | 0.790838 | 3 | 0 | 3 |
| HOXC9 | POC1A | 0.784876 | 3 | 0 | 3 |
| HOXC9 | RELB | 0.781261 | 5 | 0 | 5 |
| HOXC9 | HIVEP2 | 0.773306 | 3 | 0 | 3 |
| HOXC9 | SERPINH1 | 0.768438 | 4 | 0 | 4 |
| HOXC9 | YEATS2 | 0.766909 | 5 | 0 | 4 |
| HOXC9 | RAD54B | 0.766528 | 4 | 0 | 4 |
| HOXC9 | COLGALT1 | 0.766404 | 4 | 0 | 4 |
| HOXC9 | PHF12 | 0.765367 | 4 | 0 | 4 |
| HOXC9 | STRIP2 | 0.76169 | 3 | 0 | 3 |
| HOXC9 | CCNJL | 0.759269 | 3 | 0 | 3 |
| HOXC9 | TBC1D31 | 0.758269 | 4 | 0 | 4 |
| HOXC9 | BEND3 | 0.755652 | 6 | 0 | 6 |
| HOXC9 | NELL2 | 0.75552 | 4 | 0 | 4 |
For details and further investigation, click here