| Full name: HRas proto-oncogene, GTPase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11p15.5 | ||
| Entrez ID: 3265 | HGNC ID: HGNC:5173 | Ensembl Gene: ENSG00000174775 | OMIM ID: 190020 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
HRAS involved pathways:
Expression of HRAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HRAS | 3265 | 212983_at | 0.0425 | 0.9471 | |
| GSE20347 | HRAS | 3265 | 212983_at | -0.4223 | 0.0068 | |
| GSE23400 | HRAS | 3265 | 212983_at | -0.0743 | 0.5371 | |
| GSE26886 | HRAS | 3265 | 212983_at | -0.3943 | 0.0381 | |
| GSE29001 | HRAS | 3265 | 212983_at | -0.3463 | 0.3440 | |
| GSE38129 | HRAS | 3265 | 212983_at | -0.1055 | 0.5212 | |
| GSE45670 | HRAS | 3265 | 212983_at | 0.3061 | 0.0660 | |
| GSE53622 | HRAS | 3265 | 8197 | -0.1132 | 0.2434 | |
| GSE53624 | HRAS | 3265 | 8197 | -0.1014 | 0.1404 | |
| GSE63941 | HRAS | 3265 | 212983_at | 0.2505 | 0.5941 | |
| GSE77861 | HRAS | 3265 | 212983_at | 0.0234 | 0.9475 | |
| GSE97050 | HRAS | 3265 | A_23_P98183 | -0.0847 | 0.7555 | |
| SRP007169 | HRAS | 3265 | RNAseq | -0.6947 | 0.0310 | |
| SRP008496 | HRAS | 3265 | RNAseq | -0.4685 | 0.0307 | |
| SRP064894 | HRAS | 3265 | RNAseq | 0.5284 | 0.1222 | |
| SRP133303 | HRAS | 3265 | RNAseq | 0.0003 | 0.9985 | |
| SRP159526 | HRAS | 3265 | RNAseq | -0.3235 | 0.3511 | |
| SRP193095 | HRAS | 3265 | RNAseq | -0.2204 | 0.2731 | |
| SRP219564 | HRAS | 3265 | RNAseq | 0.0005 | 0.9991 | |
| TCGA | HRAS | 3265 | RNAseq | 0.3336 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
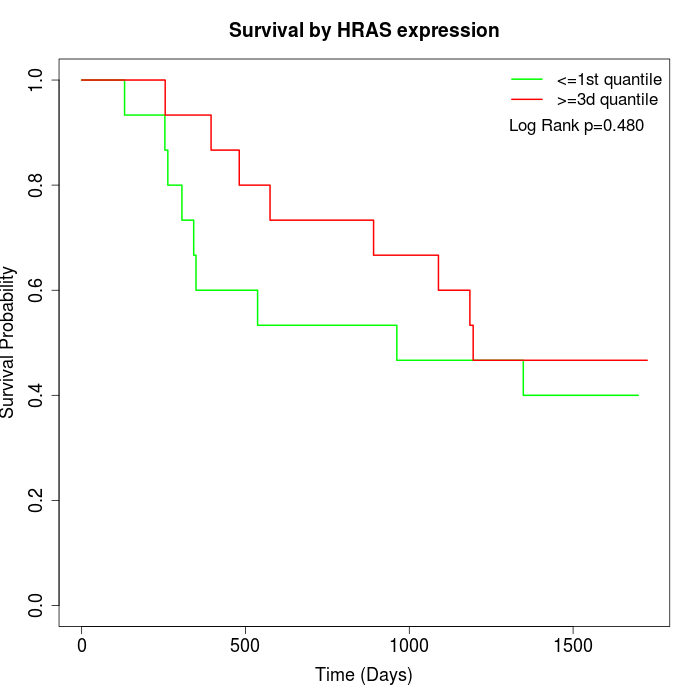
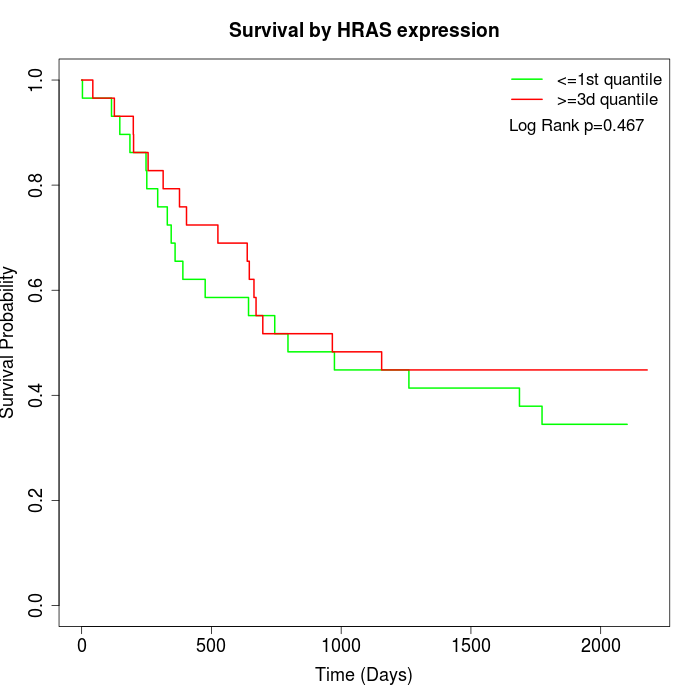
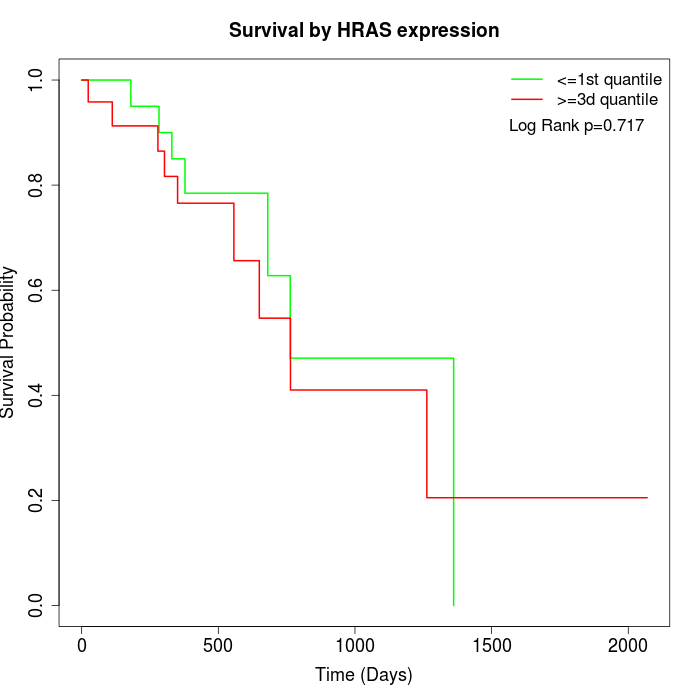
Survival by HRAS expression:
Note: Click image to view full size file.
Copy number change of HRAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HRAS | 3265 | 1 | 11 | 18 | |
| GSE20123 | HRAS | 3265 | 1 | 12 | 17 | |
| GSE43470 | HRAS | 3265 | 1 | 10 | 32 | |
| GSE46452 | HRAS | 3265 | 7 | 8 | 44 | |
| GSE47630 | HRAS | 3265 | 4 | 12 | 24 | |
| GSE54993 | HRAS | 3265 | 3 | 2 | 65 | |
| GSE54994 | HRAS | 3265 | 1 | 12 | 40 | |
| GSE60625 | HRAS | 3265 | 0 | 0 | 11 | |
| GSE74703 | HRAS | 3265 | 1 | 8 | 27 | |
| GSE74704 | HRAS | 3265 | 0 | 8 | 12 | |
| TCGA | HRAS | 3265 | 8 | 38 | 50 |
Total number of gains: 27; Total number of losses: 121; Total Number of normals: 340.
Somatic mutations of HRAS:
Generating mutation plots.
Highly correlated genes for HRAS:
Showing top 20/198 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HRAS | NHSL1 | 0.663418 | 3 | 0 | 3 |
| HRAS | FDXR | 0.634117 | 4 | 0 | 4 |
| HRAS | PGLYRP3 | 0.634106 | 3 | 0 | 3 |
| HRAS | MRPL23 | 0.627367 | 11 | 0 | 8 |
| HRAS | RUSC1 | 0.622013 | 6 | 0 | 4 |
| HRAS | RNF6 | 0.617255 | 5 | 0 | 5 |
| HRAS | ZDHHC12 | 0.605664 | 6 | 0 | 5 |
| HRAS | PKP3 | 0.603288 | 11 | 0 | 8 |
| HRAS | TALDO1 | 0.602254 | 7 | 0 | 5 |
| HRAS | AGAP3 | 0.592897 | 4 | 0 | 3 |
| HRAS | KRT5 | 0.590697 | 10 | 0 | 8 |
| HRAS | ZNF385A | 0.589433 | 3 | 0 | 3 |
| HRAS | FAM83G | 0.589224 | 6 | 0 | 4 |
| HRAS | HM13 | 0.5875 | 5 | 0 | 5 |
| HRAS | DGKA | 0.581938 | 10 | 0 | 6 |
| HRAS | LAD1 | 0.581713 | 11 | 0 | 9 |
| HRAS | RNH1 | 0.580951 | 9 | 0 | 7 |
| HRAS | GSTP1 | 0.577312 | 9 | 0 | 8 |
| HRAS | PGAP2 | 0.576324 | 8 | 0 | 6 |
| HRAS | ESPN | 0.576044 | 5 | 0 | 3 |
For details and further investigation, click here