| Full name: hyaluronidase 2 | Alias Symbol: LuCa-2|LUCA2 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 8692 | HGNC ID: HGNC:5321 | Ensembl Gene: ENSG00000068001 | OMIM ID: 603551 |
| Drug and gene relationship at DGIdb | |||
Expression of HYAL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HYAL2 | 8692 | 206855_s_at | 0.1489 | 0.6436 | |
| GSE20347 | HYAL2 | 8692 | 206855_s_at | -0.1894 | 0.0137 | |
| GSE23400 | HYAL2 | 8692 | 206855_s_at | -0.0459 | 0.3058 | |
| GSE26886 | HYAL2 | 8692 | 206855_s_at | -0.1578 | 0.3691 | |
| GSE29001 | HYAL2 | 8692 | 206855_s_at | -0.0191 | 0.9595 | |
| GSE38129 | HYAL2 | 8692 | 206855_s_at | -0.1564 | 0.0544 | |
| GSE45670 | HYAL2 | 8692 | 206855_s_at | -0.0740 | 0.6124 | |
| GSE53622 | HYAL2 | 8692 | 89478 | -0.3459 | 0.0000 | |
| GSE53624 | HYAL2 | 8692 | 89478 | -0.2761 | 0.0000 | |
| GSE63941 | HYAL2 | 8692 | 206855_s_at | -0.8169 | 0.0028 | |
| GSE77861 | HYAL2 | 8692 | 206855_s_at | -0.0060 | 0.9761 | |
| GSE97050 | HYAL2 | 8692 | A_33_P3365357 | 0.4133 | 0.2171 | |
| SRP007169 | HYAL2 | 8692 | RNAseq | 0.1246 | 0.7506 | |
| SRP008496 | HYAL2 | 8692 | RNAseq | 0.3203 | 0.1353 | |
| SRP064894 | HYAL2 | 8692 | RNAseq | 0.6870 | 0.0070 | |
| SRP133303 | HYAL2 | 8692 | RNAseq | -0.0665 | 0.7129 | |
| SRP159526 | HYAL2 | 8692 | RNAseq | -0.0453 | 0.8846 | |
| SRP193095 | HYAL2 | 8692 | RNAseq | 0.0710 | 0.6291 | |
| SRP219564 | HYAL2 | 8692 | RNAseq | -0.0559 | 0.8850 | |
| TCGA | HYAL2 | 8692 | RNAseq | -0.1968 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
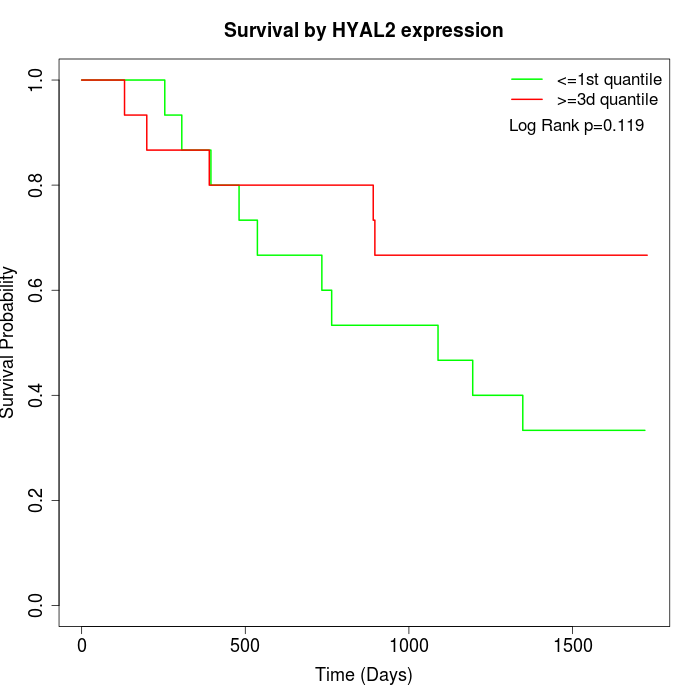
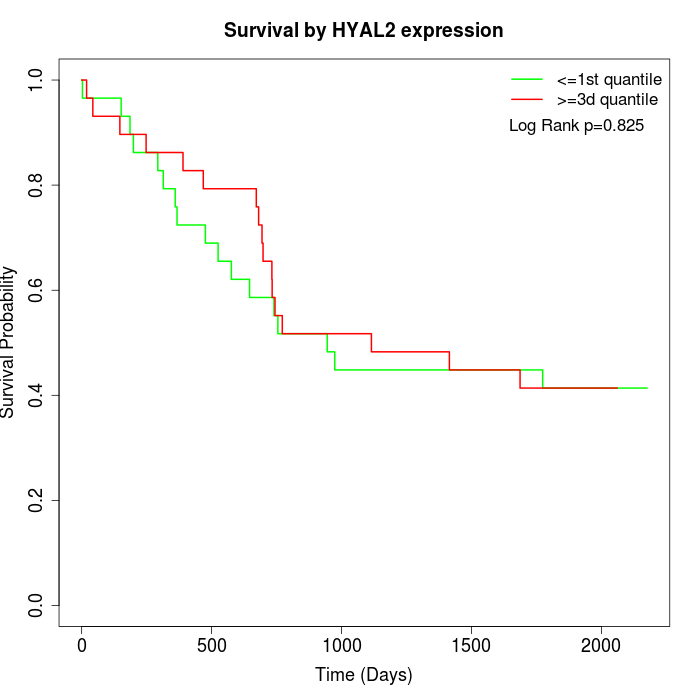
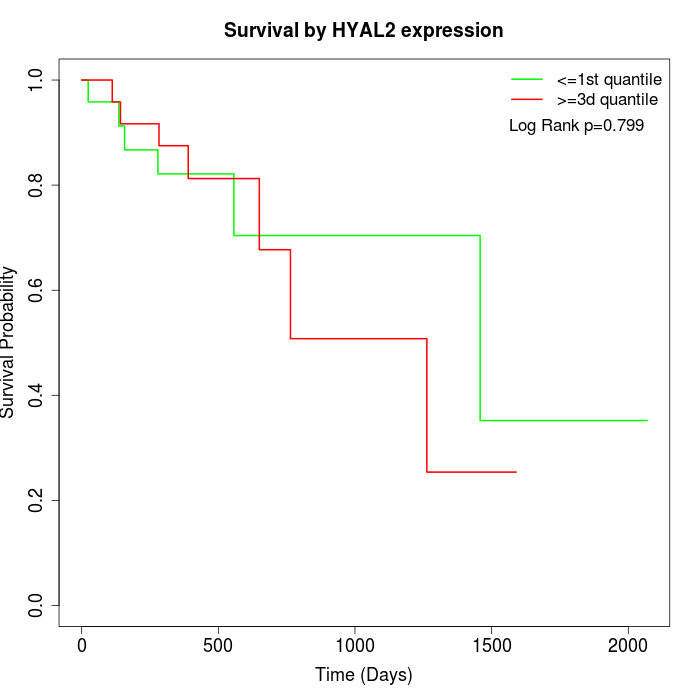
Survival by HYAL2 expression:
Note: Click image to view full size file.
Copy number change of HYAL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HYAL2 | 8692 | 0 | 18 | 12 | |
| GSE20123 | HYAL2 | 8692 | 0 | 19 | 11 | |
| GSE43470 | HYAL2 | 8692 | 0 | 19 | 24 | |
| GSE46452 | HYAL2 | 8692 | 2 | 16 | 41 | |
| GSE47630 | HYAL2 | 8692 | 1 | 24 | 15 | |
| GSE54993 | HYAL2 | 8692 | 6 | 2 | 62 | |
| GSE54994 | HYAL2 | 8692 | 1 | 33 | 19 | |
| GSE60625 | HYAL2 | 8692 | 5 | 0 | 6 | |
| GSE74703 | HYAL2 | 8692 | 0 | 15 | 21 | |
| GSE74704 | HYAL2 | 8692 | 0 | 12 | 8 | |
| TCGA | HYAL2 | 8692 | 0 | 78 | 18 |
Total number of gains: 15; Total number of losses: 236; Total Number of normals: 237.
Somatic mutations of HYAL2:
Generating mutation plots.
Highly correlated genes for HYAL2:
Showing top 20/114 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HYAL2 | CORO1B | 0.750842 | 3 | 0 | 3 |
| HYAL2 | TRPA1 | 0.695622 | 3 | 0 | 3 |
| HYAL2 | TMEM119 | 0.674537 | 3 | 0 | 3 |
| HYAL2 | SPOCD1 | 0.668378 | 3 | 0 | 3 |
| HYAL2 | PITPNM1 | 0.658659 | 3 | 0 | 3 |
| HYAL2 | MEIS3 | 0.653991 | 3 | 0 | 3 |
| HYAL2 | SPHK1 | 0.64848 | 3 | 0 | 3 |
| HYAL2 | SATB2 | 0.635977 | 3 | 0 | 3 |
| HYAL2 | SETD7 | 0.631691 | 3 | 0 | 3 |
| HYAL2 | TUBA1C | 0.629607 | 3 | 0 | 3 |
| HYAL2 | NOX4 | 0.627942 | 4 | 0 | 3 |
| HYAL2 | IL11 | 0.624777 | 4 | 0 | 3 |
| HYAL2 | CNRIP1 | 0.621827 | 4 | 0 | 4 |
| HYAL2 | ZDHHC24 | 0.619795 | 3 | 0 | 3 |
| HYAL2 | C12orf75 | 0.617162 | 3 | 0 | 3 |
| HYAL2 | MICAL1 | 0.614947 | 5 | 0 | 3 |
| HYAL2 | PHLDA3 | 0.60579 | 3 | 0 | 3 |
| HYAL2 | APLNR | 0.604674 | 7 | 0 | 6 |
| HYAL2 | SRL | 0.60227 | 3 | 0 | 3 |
| HYAL2 | TIE1 | 0.597296 | 7 | 0 | 7 |
For details and further investigation, click here