| Full name: interferon alpha and beta receptor subunit 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 21q22.11 | ||
| Entrez ID: 3455 | HGNC ID: HGNC:5433 | Ensembl Gene: ENSG00000159110 | OMIM ID: 602376 |
| Drug and gene relationship at DGIdb | |||
IFNAR2 involved pathways:
Expression of IFNAR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IFNAR2 | 3455 | 204785_x_at | 0.3803 | 0.3042 | |
| GSE20347 | IFNAR2 | 3455 | 204786_s_at | 0.6087 | 0.0005 | |
| GSE23400 | IFNAR2 | 3455 | 204785_x_at | 0.2066 | 0.0000 | |
| GSE26886 | IFNAR2 | 3455 | 204785_x_at | 0.5283 | 0.0259 | |
| GSE29001 | IFNAR2 | 3455 | 204785_x_at | 0.2358 | 0.1835 | |
| GSE38129 | IFNAR2 | 3455 | 204785_x_at | 0.2230 | 0.0808 | |
| GSE45670 | IFNAR2 | 3455 | 204785_x_at | 0.2495 | 0.0352 | |
| GSE53622 | IFNAR2 | 3455 | 16354 | 0.3883 | 0.0000 | |
| GSE53624 | IFNAR2 | 3455 | 16354 | 0.5746 | 0.0000 | |
| GSE63941 | IFNAR2 | 3455 | 204785_x_at | 0.2299 | 0.6494 | |
| GSE77861 | IFNAR2 | 3455 | 204786_s_at | 0.1477 | 0.3384 | |
| GSE97050 | IFNAR2 | 3455 | A_33_P3361457 | 0.7985 | 0.1330 | |
| SRP007169 | IFNAR2 | 3455 | RNAseq | 0.7097 | 0.1513 | |
| SRP008496 | IFNAR2 | 3455 | RNAseq | 0.1428 | 0.6957 | |
| SRP064894 | IFNAR2 | 3455 | RNAseq | 0.7706 | 0.0000 | |
| SRP133303 | IFNAR2 | 3455 | RNAseq | 0.4008 | 0.0133 | |
| SRP159526 | IFNAR2 | 3455 | RNAseq | 0.3869 | 0.1980 | |
| SRP193095 | IFNAR2 | 3455 | RNAseq | 0.4560 | 0.0072 | |
| SRP219564 | IFNAR2 | 3455 | RNAseq | 0.3025 | 0.2587 | |
| TCGA | IFNAR2 | 3455 | RNAseq | 0.0456 | 0.4840 |
Upregulated datasets: 0; Downregulated datasets: 0.
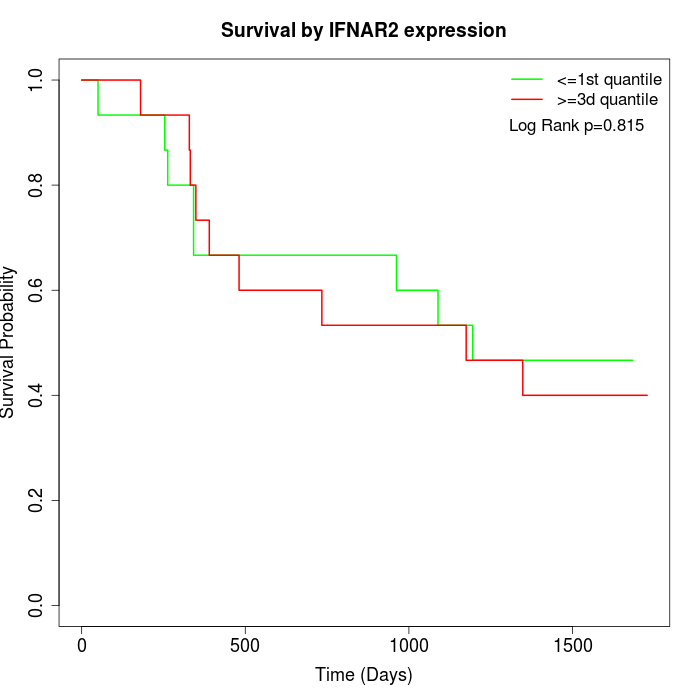
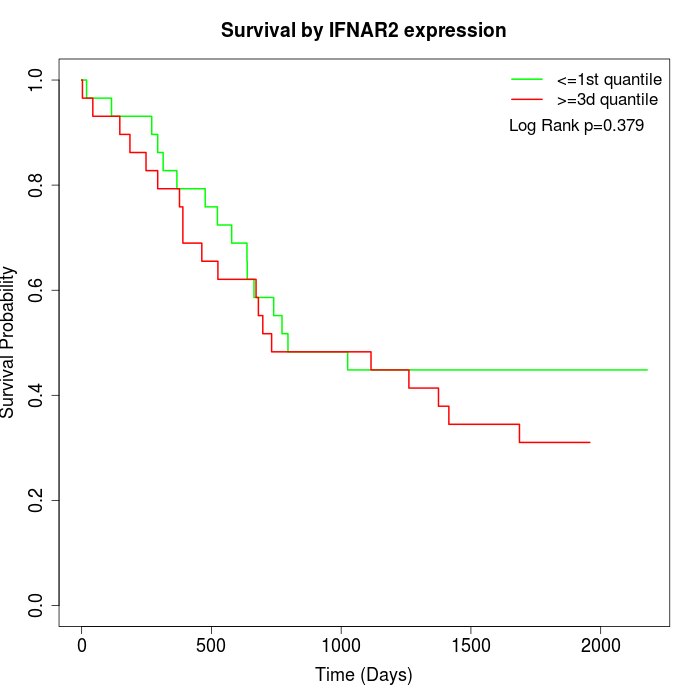
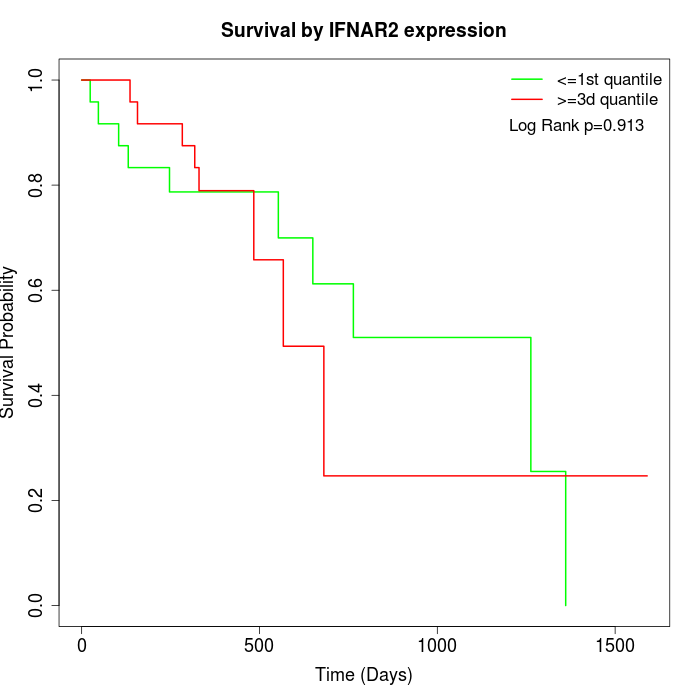
Survival by IFNAR2 expression:
Note: Click image to view full size file.
Copy number change of IFNAR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IFNAR2 | 3455 | 2 | 8 | 20 | |
| GSE20123 | IFNAR2 | 3455 | 2 | 9 | 19 | |
| GSE43470 | IFNAR2 | 3455 | 3 | 9 | 31 | |
| GSE46452 | IFNAR2 | 3455 | 1 | 21 | 37 | |
| GSE47630 | IFNAR2 | 3455 | 6 | 17 | 17 | |
| GSE54993 | IFNAR2 | 3455 | 9 | 1 | 60 | |
| GSE54994 | IFNAR2 | 3455 | 1 | 9 | 43 | |
| GSE60625 | IFNAR2 | 3455 | 0 | 0 | 11 | |
| GSE74703 | IFNAR2 | 3455 | 3 | 6 | 27 | |
| GSE74704 | IFNAR2 | 3455 | 1 | 4 | 15 | |
| TCGA | IFNAR2 | 3455 | 9 | 35 | 52 |
Total number of gains: 37; Total number of losses: 119; Total Number of normals: 332.
Somatic mutations of IFNAR2:
Generating mutation plots.
Highly correlated genes for IFNAR2:
Showing top 20/671 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IFNAR2 | CCNK | 0.733231 | 3 | 0 | 3 |
| IFNAR2 | PDIA5 | 0.726123 | 3 | 0 | 3 |
| IFNAR2 | PSMD13 | 0.689917 | 3 | 0 | 3 |
| IFNAR2 | AFAP1L2 | 0.68928 | 3 | 0 | 3 |
| IFNAR2 | C1orf35 | 0.683404 | 3 | 0 | 3 |
| IFNAR2 | PREX1 | 0.678271 | 4 | 0 | 4 |
| IFNAR2 | NPLOC4 | 0.675964 | 4 | 0 | 3 |
| IFNAR2 | LRRC8C | 0.67061 | 3 | 0 | 3 |
| IFNAR2 | LINC00597 | 0.666064 | 3 | 0 | 3 |
| IFNAR2 | VAV1 | 0.664259 | 4 | 0 | 4 |
| IFNAR2 | LMO1 | 0.660278 | 3 | 0 | 3 |
| IFNAR2 | EPSTI1 | 0.658778 | 4 | 0 | 4 |
| IFNAR2 | ZBED4 | 0.656705 | 3 | 0 | 3 |
| IFNAR2 | ICAM1 | 0.655037 | 6 | 0 | 5 |
| IFNAR2 | TBXAS1 | 0.653708 | 3 | 0 | 3 |
| IFNAR2 | SPI1 | 0.651561 | 6 | 0 | 5 |
| IFNAR2 | BATF3 | 0.651505 | 7 | 0 | 6 |
| IFNAR2 | PIK3AP1 | 0.651189 | 5 | 0 | 4 |
| IFNAR2 | TTC39C | 0.65089 | 3 | 0 | 3 |
| IFNAR2 | CTSZ | 0.648993 | 5 | 0 | 4 |
For details and further investigation, click here