| Full name: integrin subunit alpha M | Alias Symbol: MAC-1|CD11b | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 3684 | HGNC ID: HGNC:6149 | Ensembl Gene: ENSG00000169896 | OMIM ID: 120980 |
| Drug and gene relationship at DGIdb | |||
ITGAM involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05140 | Leishmaniasis | |
| hsa05152 | Tuberculosis |
Expression of ITGAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGAM | 3684 | 205786_s_at | 0.2581 | 0.8684 | |
| GSE26886 | ITGAM | 3684 | 205786_s_at | 1.0141 | 0.0144 | |
| GSE29001 | ITGAM | 3684 | 205786_s_at | 0.6862 | 0.0329 | |
| GSE38129 | ITGAM | 3684 | 205786_s_at | 0.1876 | 0.3693 | |
| GSE45670 | ITGAM | 3684 | 205786_s_at | -0.5824 | 0.1617 | |
| GSE53622 | ITGAM | 3684 | 151178 | 0.4777 | 0.0148 | |
| GSE53624 | ITGAM | 3684 | 151178 | 0.4517 | 0.0457 | |
| GSE97050 | ITGAM | 3684 | A_23_P124108 | 0.6125 | 0.1195 | |
| SRP007169 | ITGAM | 3684 | RNAseq | 3.7356 | 0.0001 | |
| SRP008496 | ITGAM | 3684 | RNAseq | 3.7616 | 0.0000 | |
| SRP064894 | ITGAM | 3684 | RNAseq | 1.1910 | 0.0039 | |
| SRP133303 | ITGAM | 3684 | RNAseq | 0.5532 | 0.0308 | |
| SRP159526 | ITGAM | 3684 | RNAseq | 0.6213 | 0.2553 | |
| SRP193095 | ITGAM | 3684 | RNAseq | 1.2396 | 0.0000 | |
| SRP219564 | ITGAM | 3684 | RNAseq | 1.4848 | 0.0505 | |
| TCGA | ITGAM | 3684 | RNAseq | 0.2817 | 0.1362 |
Upregulated datasets: 5; Downregulated datasets: 0.
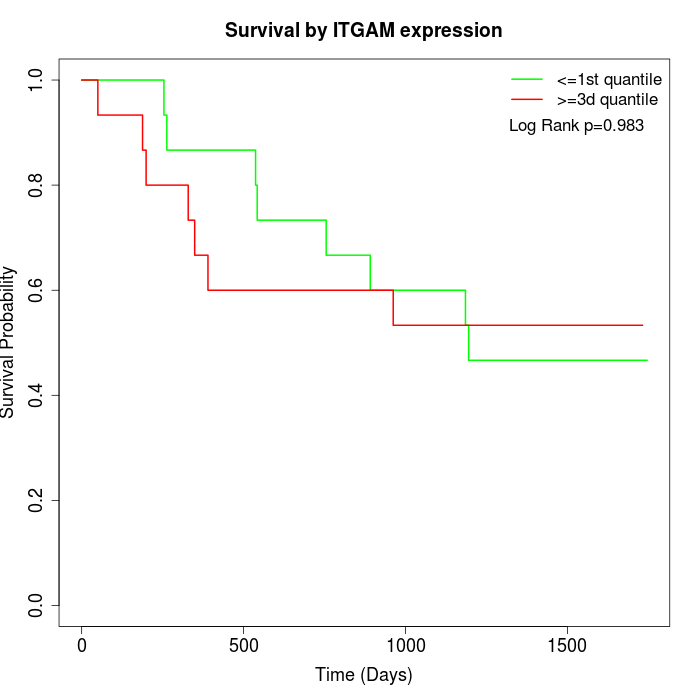
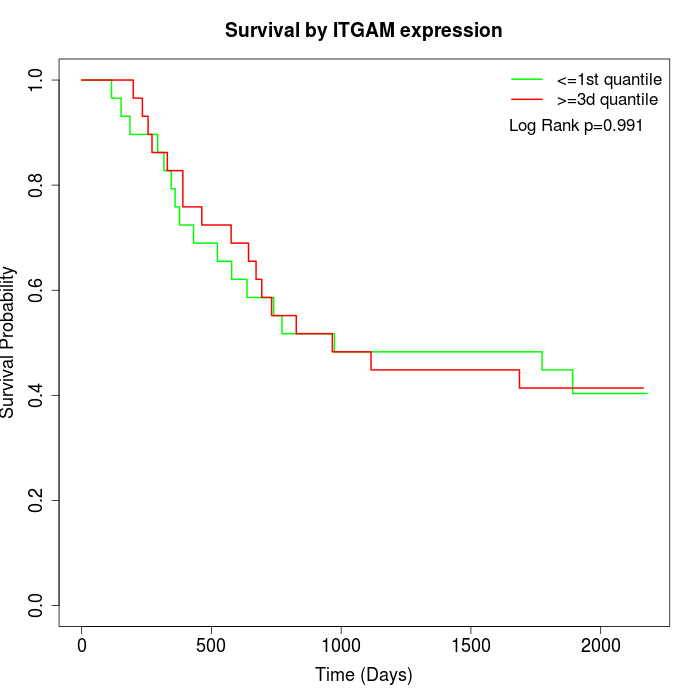
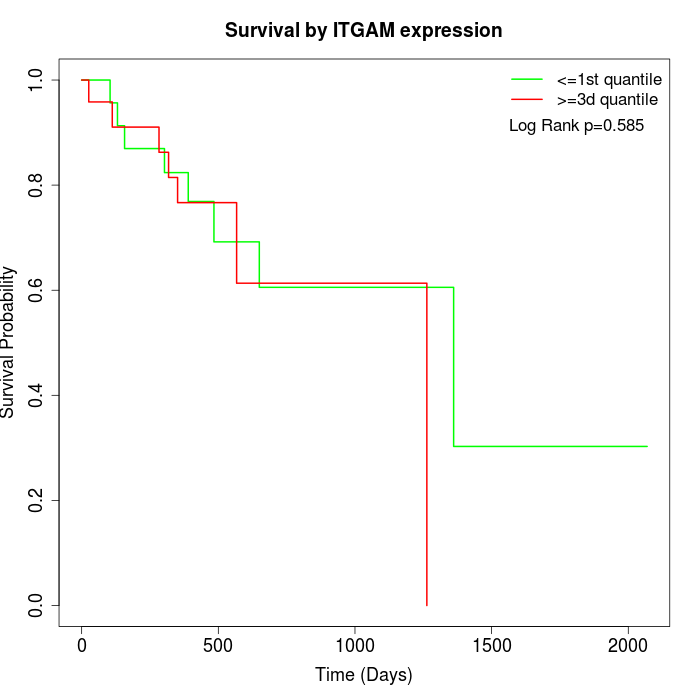
Survival by ITGAM expression:
Note: Click image to view full size file.
Copy number change of ITGAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGAM | 3684 | 4 | 5 | 21 | |
| GSE20123 | ITGAM | 3684 | 4 | 4 | 22 | |
| GSE43470 | ITGAM | 3684 | 3 | 3 | 37 | |
| GSE46452 | ITGAM | 3684 | 38 | 1 | 20 | |
| GSE47630 | ITGAM | 3684 | 11 | 7 | 22 | |
| GSE54993 | ITGAM | 3684 | 2 | 5 | 63 | |
| GSE54994 | ITGAM | 3684 | 5 | 9 | 39 | |
| GSE60625 | ITGAM | 3684 | 4 | 0 | 7 | |
| GSE74703 | ITGAM | 3684 | 3 | 2 | 31 | |
| GSE74704 | ITGAM | 3684 | 2 | 2 | 16 | |
| TCGA | ITGAM | 3684 | 20 | 10 | 66 |
Total number of gains: 96; Total number of losses: 48; Total Number of normals: 344.
Somatic mutations of ITGAM:
Generating mutation plots.
Highly correlated genes for ITGAM:
Showing top 20/949 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGAM | C19orf38 | 0.80737 | 3 | 0 | 3 |
| ITGAM | GNGT2 | 0.796036 | 3 | 0 | 3 |
| ITGAM | RAB39B | 0.764662 | 3 | 0 | 3 |
| ITGAM | FCGR3A | 0.764323 | 3 | 0 | 3 |
| ITGAM | GPR141 | 0.757434 | 3 | 0 | 3 |
| ITGAM | SUCNR1 | 0.756616 | 4 | 0 | 4 |
| ITGAM | CD163 | 0.750335 | 9 | 0 | 9 |
| ITGAM | CD84 | 0.741309 | 7 | 0 | 7 |
| ITGAM | SH2B3 | 0.734832 | 9 | 0 | 8 |
| ITGAM | SRGN | 0.733954 | 9 | 0 | 9 |
| ITGAM | CYBB | 0.733887 | 8 | 0 | 8 |
| ITGAM | IL2RA | 0.732668 | 4 | 0 | 4 |
| ITGAM | EBI3 | 0.732632 | 4 | 0 | 4 |
| ITGAM | LRRK1 | 0.730814 | 3 | 0 | 3 |
| ITGAM | JTB | 0.727837 | 3 | 0 | 3 |
| ITGAM | CERKL | 0.725995 | 3 | 0 | 3 |
| ITGAM | TMEM176B | 0.725542 | 9 | 0 | 8 |
| ITGAM | CD180 | 0.724325 | 5 | 0 | 5 |
| ITGAM | CKAP5 | 0.722107 | 3 | 0 | 3 |
| ITGAM | GMPS | 0.721149 | 3 | 0 | 3 |
For details and further investigation, click here