| Full name: potassium voltage-gated channel subfamily C member 1 | Alias Symbol: Kv3.1 | ||
| Type: protein-coding gene | Cytoband: 11p15.1 | ||
| Entrez ID: 3746 | HGNC ID: HGNC:6233 | Ensembl Gene: ENSG00000129159 | OMIM ID: 176258 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNC1 | 3746 | 208477_at | -0.1771 | 0.5190 | |
| GSE20347 | KCNC1 | 3746 | 208477_at | -0.0381 | 0.6243 | |
| GSE23400 | KCNC1 | 3746 | 208477_at | -0.1197 | 0.0102 | |
| GSE26886 | KCNC1 | 3746 | 208477_at | 0.0886 | 0.3992 | |
| GSE29001 | KCNC1 | 3746 | 208477_at | -0.1607 | 0.4068 | |
| GSE38129 | KCNC1 | 3746 | 208477_at | -0.0589 | 0.3816 | |
| GSE45670 | KCNC1 | 3746 | 208477_at | 0.0902 | 0.2289 | |
| GSE53622 | KCNC1 | 3746 | 99217 | 0.4219 | 0.0513 | |
| GSE53624 | KCNC1 | 3746 | 99217 | -0.0194 | 0.9248 | |
| GSE63941 | KCNC1 | 3746 | 208477_at | 0.2465 | 0.0430 | |
| GSE77861 | KCNC1 | 3746 | 208477_at | -0.0577 | 0.6569 | |
| TCGA | KCNC1 | 3746 | RNAseq | -0.6961 | 0.2229 |
Upregulated datasets: 0; Downregulated datasets: 0.
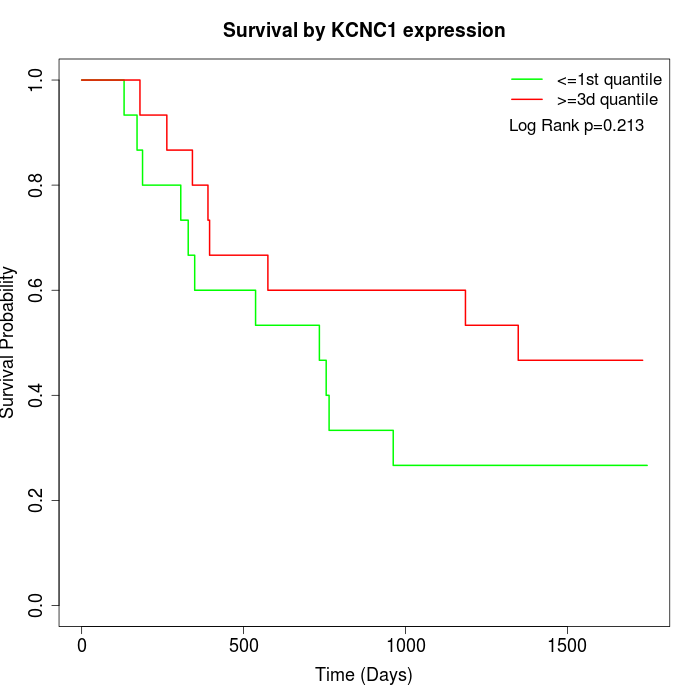
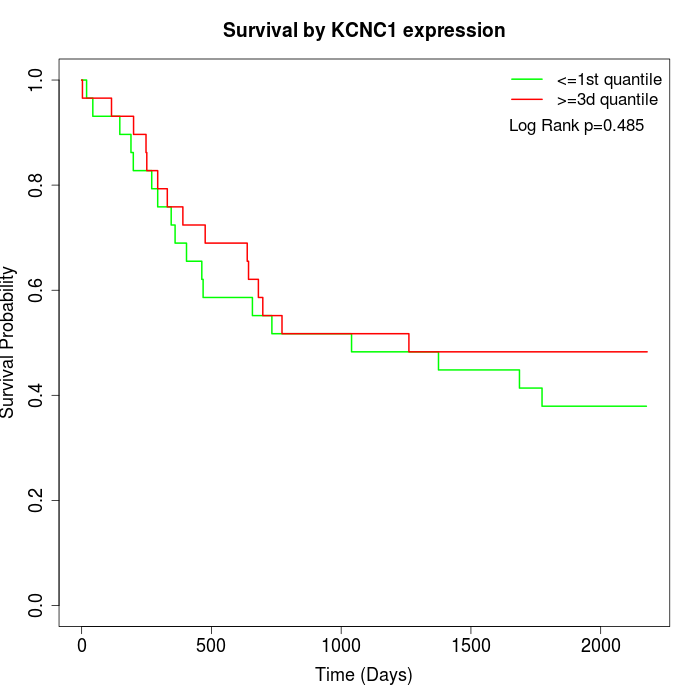
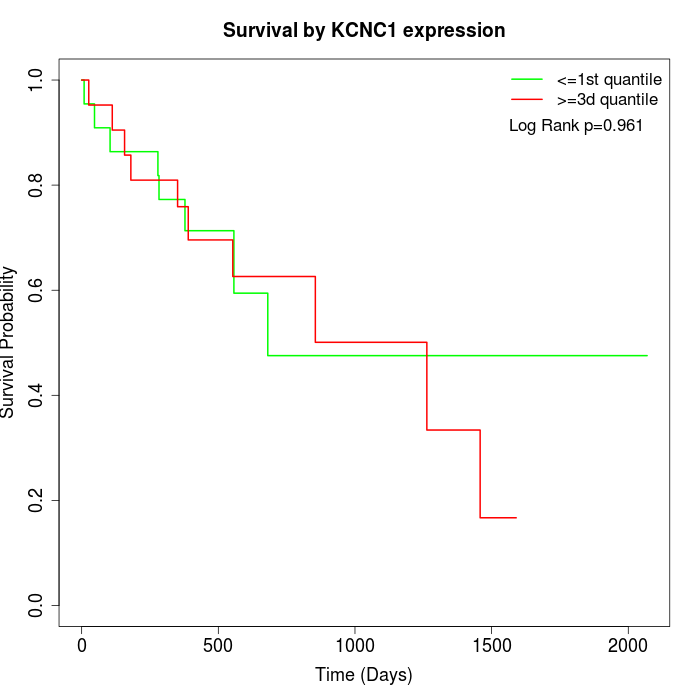
Survival by KCNC1 expression:
Note: Click image to view full size file.
Copy number change of KCNC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNC1 | 3746 | 0 | 10 | 20 | |
| GSE20123 | KCNC1 | 3746 | 0 | 10 | 20 | |
| GSE43470 | KCNC1 | 3746 | 1 | 5 | 37 | |
| GSE46452 | KCNC1 | 3746 | 7 | 5 | 47 | |
| GSE47630 | KCNC1 | 3746 | 3 | 10 | 27 | |
| GSE54993 | KCNC1 | 3746 | 3 | 0 | 67 | |
| GSE54994 | KCNC1 | 3746 | 3 | 12 | 38 | |
| GSE60625 | KCNC1 | 3746 | 0 | 0 | 11 | |
| GSE74703 | KCNC1 | 3746 | 1 | 3 | 32 | |
| GSE74704 | KCNC1 | 3746 | 0 | 9 | 11 | |
| TCGA | KCNC1 | 3746 | 11 | 29 | 56 |
Total number of gains: 29; Total number of losses: 93; Total Number of normals: 366.
Somatic mutations of KCNC1:
Generating mutation plots.
Highly correlated genes for KCNC1:
Showing top 20/806 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNC1 | GFRA3 | 0.737199 | 3 | 0 | 3 |
| KCNC1 | PSG9 | 0.703279 | 3 | 0 | 3 |
| KCNC1 | APOBEC1 | 0.68869 | 3 | 0 | 3 |
| KCNC1 | GRIN1 | 0.686254 | 4 | 0 | 4 |
| KCNC1 | KRTAP9-9 | 0.679465 | 3 | 0 | 3 |
| KCNC1 | ZNF81 | 0.679381 | 3 | 0 | 3 |
| KCNC1 | SLC18A1 | 0.674072 | 4 | 0 | 4 |
| KCNC1 | GRIA1 | 0.668186 | 3 | 0 | 3 |
| KCNC1 | CACTIN | 0.667893 | 3 | 0 | 3 |
| KCNC1 | OR3A1 | 0.666598 | 3 | 0 | 3 |
| KCNC1 | RPS6KA6 | 0.666583 | 5 | 0 | 4 |
| KCNC1 | ITIH2 | 0.666558 | 7 | 0 | 5 |
| KCNC1 | ADAM21 | 0.665515 | 3 | 0 | 3 |
| KCNC1 | IL1RL1 | 0.66222 | 3 | 0 | 3 |
| KCNC1 | PRB4 | 0.661061 | 8 | 0 | 8 |
| KCNC1 | CA14 | 0.660914 | 6 | 0 | 6 |
| KCNC1 | RABEP2 | 0.660574 | 4 | 0 | 4 |
| KCNC1 | GIP | 0.6595 | 4 | 0 | 3 |
| KCNC1 | MTTP | 0.657833 | 4 | 0 | 3 |
| KCNC1 | FSCN3 | 0.656758 | 5 | 0 | 4 |
For details and further investigation, click here