| Full name: kelch like family member 25 | Alias Symbol: FLJ12587|ENC2|ENC-2 | ||
| Type: protein-coding gene | Cytoband: 15q25.3 | ||
| Entrez ID: 64410 | HGNC ID: HGNC:25732 | Ensembl Gene: ENSG00000183655 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL25:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL25 | 64410 | 210307_s_at | -0.1320 | 0.7346 | |
| GSE20347 | KLHL25 | 64410 | 210307_s_at | 0.1171 | 0.1161 | |
| GSE23400 | KLHL25 | 64410 | 210307_s_at | -0.1086 | 0.0026 | |
| GSE26886 | KLHL25 | 64410 | 210307_s_at | 0.3754 | 0.0020 | |
| GSE29001 | KLHL25 | 64410 | 210307_s_at | 0.0036 | 0.9841 | |
| GSE38129 | KLHL25 | 64410 | 210307_s_at | -0.0107 | 0.8994 | |
| GSE45670 | KLHL25 | 64410 | 210307_s_at | 0.0938 | 0.5675 | |
| GSE53622 | KLHL25 | 64410 | 54452 | -0.2005 | 0.0314 | |
| GSE53624 | KLHL25 | 64410 | 54452 | -0.2172 | 0.0122 | |
| GSE63941 | KLHL25 | 64410 | 210307_s_at | 0.2634 | 0.3057 | |
| GSE77861 | KLHL25 | 64410 | 210307_s_at | 0.0473 | 0.6669 | |
| GSE97050 | KLHL25 | 64410 | A_23_P65797 | -0.1173 | 0.6102 | |
| SRP007169 | KLHL25 | 64410 | RNAseq | -0.3239 | 0.6079 | |
| SRP064894 | KLHL25 | 64410 | RNAseq | -0.3478 | 0.1905 | |
| SRP133303 | KLHL25 | 64410 | RNAseq | -0.1172 | 0.6700 | |
| SRP159526 | KLHL25 | 64410 | RNAseq | -0.2646 | 0.4202 | |
| SRP193095 | KLHL25 | 64410 | RNAseq | -0.3348 | 0.0146 | |
| SRP219564 | KLHL25 | 64410 | RNAseq | -0.8287 | 0.1398 | |
| TCGA | KLHL25 | 64410 | RNAseq | -0.2515 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 0.
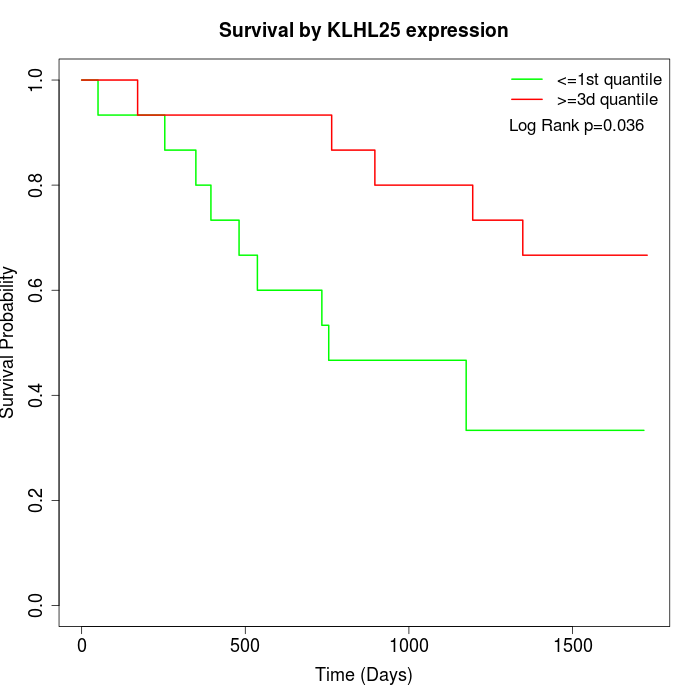
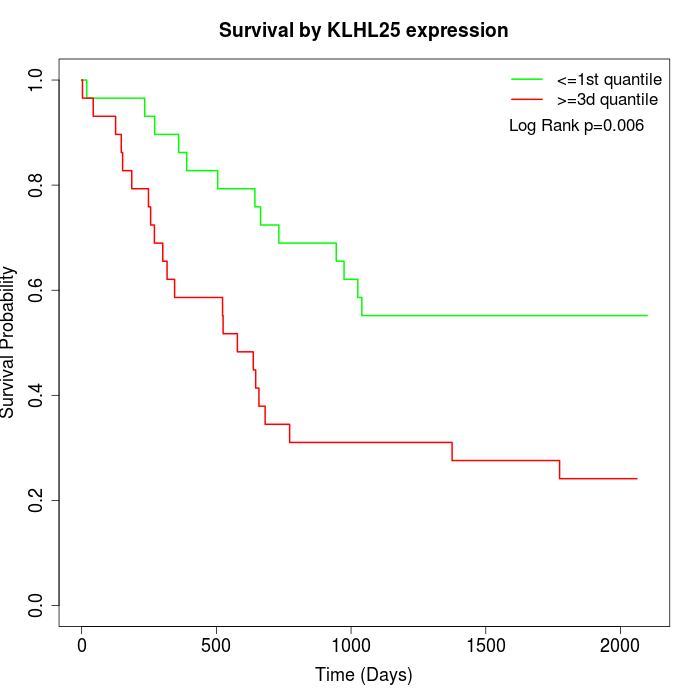
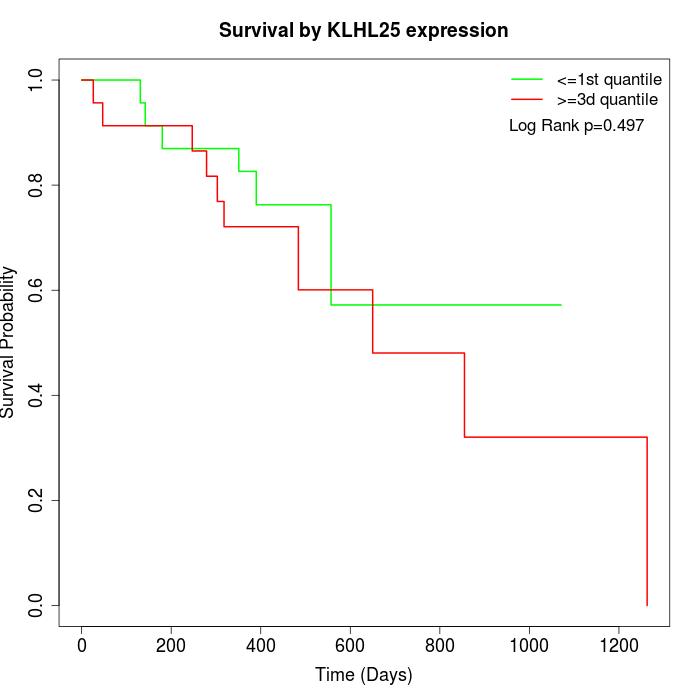
Survival by KLHL25 expression:
Note: Click image to view full size file.
Copy number change of KLHL25:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL25 | 64410 | 8 | 0 | 22 | |
| GSE20123 | KLHL25 | 64410 | 8 | 0 | 22 | |
| GSE43470 | KLHL25 | 64410 | 4 | 5 | 34 | |
| GSE46452 | KLHL25 | 64410 | 3 | 7 | 49 | |
| GSE47630 | KLHL25 | 64410 | 8 | 10 | 22 | |
| GSE54993 | KLHL25 | 64410 | 5 | 6 | 59 | |
| GSE54994 | KLHL25 | 64410 | 6 | 6 | 41 | |
| GSE60625 | KLHL25 | 64410 | 4 | 0 | 7 | |
| GSE74703 | KLHL25 | 64410 | 4 | 3 | 29 | |
| GSE74704 | KLHL25 | 64410 | 4 | 0 | 16 | |
| TCGA | KLHL25 | 64410 | 16 | 13 | 67 |
Total number of gains: 70; Total number of losses: 50; Total Number of normals: 368.
Somatic mutations of KLHL25:
Generating mutation plots.
Highly correlated genes for KLHL25:
Showing top 20/307 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL25 | CRYBA1 | 0.738297 | 3 | 0 | 3 |
| KLHL25 | FAM186B | 0.724021 | 3 | 0 | 3 |
| KLHL25 | C9orf131 | 0.707134 | 3 | 0 | 3 |
| KLHL25 | BPY2 | 0.662073 | 3 | 0 | 3 |
| KLHL25 | CHRND | 0.662031 | 3 | 0 | 3 |
| KLHL25 | JMJD6 | 0.661322 | 3 | 0 | 3 |
| KLHL25 | LIM2 | 0.656251 | 5 | 0 | 5 |
| KLHL25 | ASIC4 | 0.651817 | 5 | 0 | 5 |
| KLHL25 | EVX1 | 0.648444 | 4 | 0 | 4 |
| KLHL25 | DRD2 | 0.647228 | 4 | 0 | 4 |
| KLHL25 | MLXIPL | 0.645181 | 5 | 0 | 4 |
| KLHL25 | SOX15 | 0.642365 | 3 | 0 | 3 |
| KLHL25 | HBQ1 | 0.634969 | 5 | 0 | 4 |
| KLHL25 | MYOG | 0.6304 | 5 | 0 | 5 |
| KLHL25 | SCUBE1 | 0.629078 | 5 | 0 | 5 |
| KLHL25 | SPTB | 0.629008 | 4 | 0 | 4 |
| KLHL25 | BIRC7 | 0.62761 | 3 | 0 | 3 |
| KLHL25 | SV2A | 0.625759 | 3 | 0 | 3 |
| KLHL25 | GRK1 | 0.619334 | 5 | 0 | 4 |
| KLHL25 | FAM186A | 0.616842 | 4 | 0 | 4 |
For details and further investigation, click here