| Full name: kelch like family member 9 | Alias Symbol: KIAA1354|FLJ13568 | ||
| Type: protein-coding gene | Cytoband: 9p21.3 | ||
| Entrez ID: 55958 | HGNC ID: HGNC:18732 | Ensembl Gene: ENSG00000198642 | OMIM ID: 611201 |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL9 | 55958 | 213233_s_at | -0.9587 | 0.3328 | |
| GSE20347 | KLHL9 | 55958 | 213233_s_at | -0.6401 | 0.0037 | |
| GSE23400 | KLHL9 | 55958 | 213233_s_at | 0.0202 | 0.8286 | |
| GSE26886 | KLHL9 | 55958 | 213233_s_at | -0.1369 | 0.4566 | |
| GSE29001 | KLHL9 | 55958 | 213233_s_at | -0.3428 | 0.2934 | |
| GSE38129 | KLHL9 | 55958 | 213233_s_at | -0.3680 | 0.0190 | |
| GSE45670 | KLHL9 | 55958 | 213233_s_at | -0.2903 | 0.3142 | |
| GSE53622 | KLHL9 | 55958 | 65226 | -0.1495 | 0.1107 | |
| GSE53624 | KLHL9 | 55958 | 65226 | -0.2008 | 0.2386 | |
| GSE63941 | KLHL9 | 55958 | 213233_s_at | -1.4878 | 0.2640 | |
| GSE77861 | KLHL9 | 55958 | 213233_s_at | -0.8247 | 0.1327 | |
| GSE97050 | KLHL9 | 55958 | A_23_P83159 | -0.3985 | 0.2276 | |
| SRP007169 | KLHL9 | 55958 | RNAseq | 0.0632 | 0.8856 | |
| SRP008496 | KLHL9 | 55958 | RNAseq | 0.2592 | 0.3011 | |
| SRP064894 | KLHL9 | 55958 | RNAseq | -0.1643 | 0.3051 | |
| SRP133303 | KLHL9 | 55958 | RNAseq | -0.0950 | 0.3987 | |
| SRP159526 | KLHL9 | 55958 | RNAseq | -0.3925 | 0.1313 | |
| SRP193095 | KLHL9 | 55958 | RNAseq | -0.6293 | 0.0000 | |
| SRP219564 | KLHL9 | 55958 | RNAseq | -0.4815 | 0.0719 | |
| TCGA | KLHL9 | 55958 | RNAseq | -0.2412 | 0.0049 |
Upregulated datasets: 0; Downregulated datasets: 0.
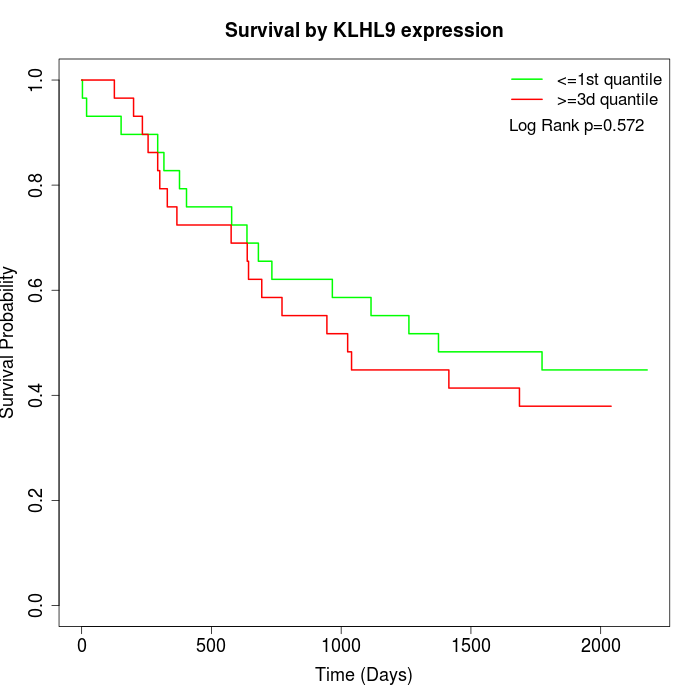
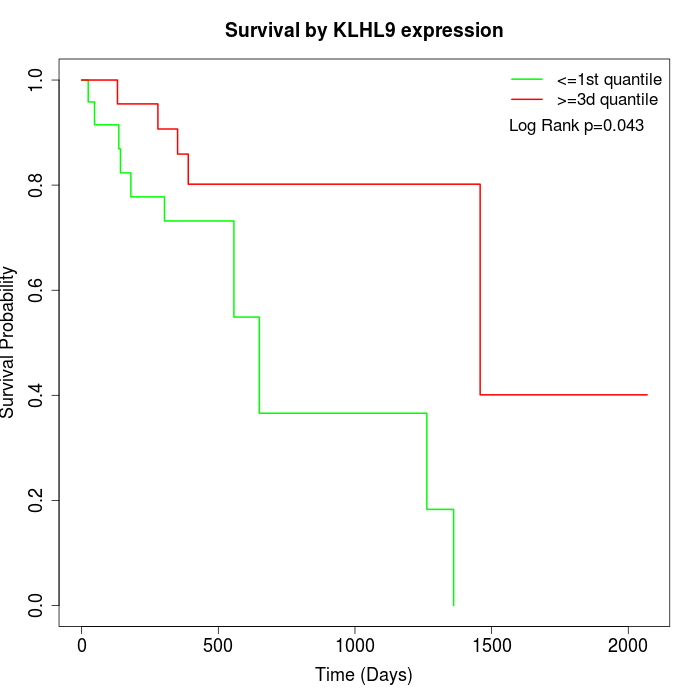
Survival by KLHL9 expression:
Note: Click image to view full size file.
Copy number change of KLHL9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL9 | 55958 | 2 | 17 | 11 | |
| GSE20123 | KLHL9 | 55958 | 2 | 17 | 11 | |
| GSE43470 | KLHL9 | 55958 | 4 | 13 | 26 | |
| GSE46452 | KLHL9 | 55958 | 2 | 24 | 33 | |
| GSE47630 | KLHL9 | 55958 | 3 | 27 | 10 | |
| GSE54993 | KLHL9 | 55958 | 7 | 2 | 61 | |
| GSE54994 | KLHL9 | 55958 | 8 | 17 | 28 | |
| GSE60625 | KLHL9 | 55958 | 3 | 0 | 8 | |
| GSE74703 | KLHL9 | 55958 | 4 | 10 | 22 | |
| GSE74704 | KLHL9 | 55958 | 0 | 13 | 7 | |
| TCGA | KLHL9 | 55958 | 11 | 53 | 32 |
Total number of gains: 46; Total number of losses: 193; Total Number of normals: 249.
Somatic mutations of KLHL9:
Generating mutation plots.
Highly correlated genes for KLHL9:
Showing top 20/427 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL9 | ZNF383 | 0.782899 | 3 | 0 | 3 |
| KLHL9 | HMGN5 | 0.782733 | 3 | 0 | 3 |
| KLHL9 | PPP3CC | 0.760166 | 3 | 0 | 3 |
| KLHL9 | NFKB1 | 0.759609 | 3 | 0 | 3 |
| KLHL9 | ACADVL | 0.758598 | 3 | 0 | 3 |
| KLHL9 | UBE2E2 | 0.753307 | 3 | 0 | 3 |
| KLHL9 | CMC1 | 0.747044 | 3 | 0 | 3 |
| KLHL9 | CGNL1 | 0.744035 | 3 | 0 | 3 |
| KLHL9 | YAP1 | 0.731659 | 3 | 0 | 3 |
| KLHL9 | TAF9B | 0.726334 | 3 | 0 | 3 |
| KLHL9 | JAK1 | 0.725416 | 3 | 0 | 3 |
| KLHL9 | OFD1 | 0.71684 | 4 | 0 | 3 |
| KLHL9 | ATG2B | 0.716244 | 3 | 0 | 3 |
| KLHL9 | CNOT6L | 0.703705 | 4 | 0 | 3 |
| KLHL9 | HDHD2 | 0.70095 | 5 | 0 | 5 |
| KLHL9 | LMBRD2 | 0.699825 | 3 | 0 | 3 |
| KLHL9 | SVIP | 0.69159 | 4 | 0 | 3 |
| KLHL9 | MTIF3 | 0.687427 | 4 | 0 | 3 |
| KLHL9 | DYM | 0.687382 | 5 | 0 | 4 |
| KLHL9 | SLC4A4 | 0.687003 | 3 | 0 | 3 |
For details and further investigation, click here