| Full name: keratin associated protein 1-3 | Alias Symbol: KAP1.3 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 81850 | HGNC ID: HGNC:16771 | Ensembl Gene: ENSG00000221880 | OMIM ID: 608820 |
| Drug and gene relationship at DGIdb | |||
Expression of KRTAP1-3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRTAP1-3 | 81850 | 234880_x_at | 0.0222 | 0.9563 | |
| GSE20347 | KRTAP1-3 | 81850 | 220978_at | -0.0432 | 0.4330 | |
| GSE23400 | KRTAP1-3 | 81850 | 220978_at | -0.0632 | 0.0049 | |
| GSE26886 | KRTAP1-3 | 81850 | 234880_x_at | 0.0938 | 0.3999 | |
| GSE29001 | KRTAP1-3 | 81850 | 220978_at | -0.0257 | 0.8568 | |
| GSE38129 | KRTAP1-3 | 81850 | 220978_at | -0.0610 | 0.1926 | |
| GSE45670 | KRTAP1-3 | 81850 | 234880_x_at | 0.0003 | 0.9984 | |
| GSE53622 | KRTAP1-3 | 81850 | 1401 | -0.2334 | 0.0002 | |
| GSE53624 | KRTAP1-3 | 81850 | 1401 | -0.2382 | 0.0024 | |
| GSE63941 | KRTAP1-3 | 81850 | 234880_x_at | -1.3213 | 0.0207 | |
| GSE77861 | KRTAP1-3 | 81850 | 234880_x_at | -0.1997 | 0.1314 | |
| GSE97050 | KRTAP1-3 | 81850 | A_33_P3213006 | -0.1459 | 0.5785 |
Upregulated datasets: 0; Downregulated datasets: 1.
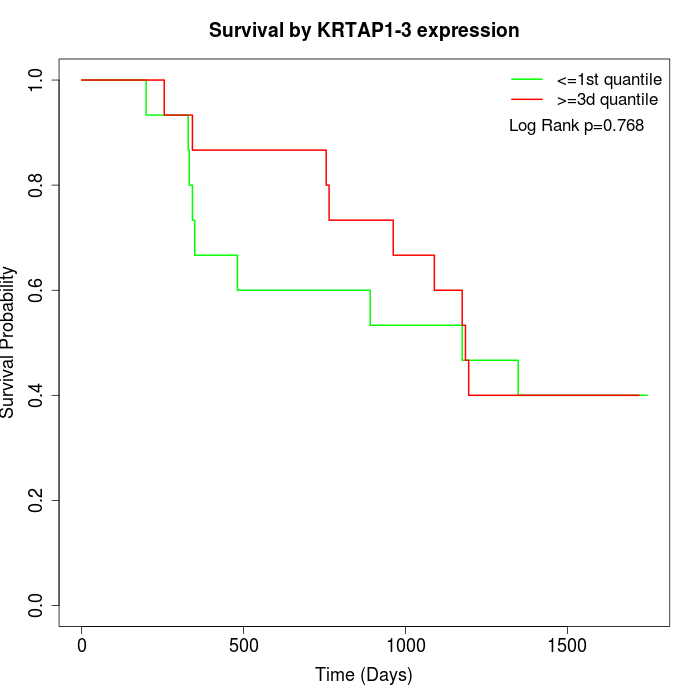
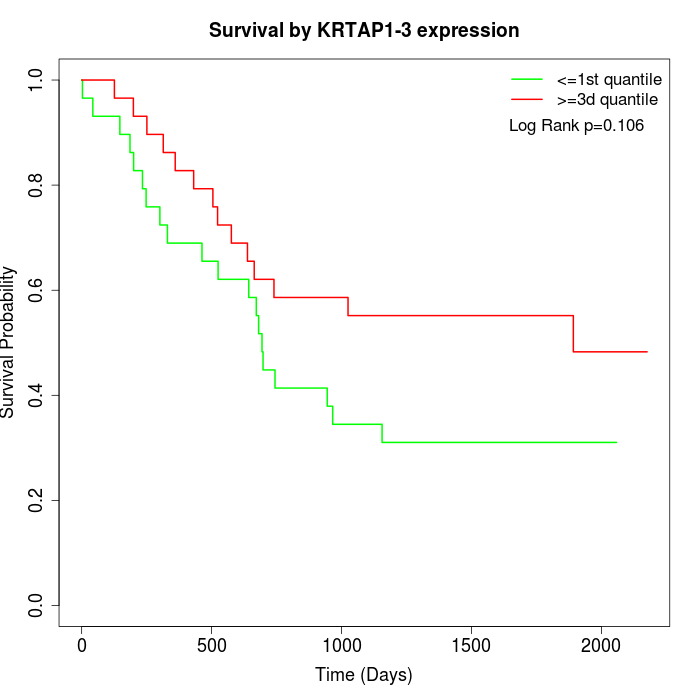
Survival by KRTAP1-3 expression:
Note: Click image to view full size file.
Copy number change of KRTAP1-3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRTAP1-3 | 81850 | 6 | 2 | 22 | |
| GSE20123 | KRTAP1-3 | 81850 | 6 | 2 | 22 | |
| GSE43470 | KRTAP1-3 | 81850 | 1 | 2 | 40 | |
| GSE46452 | KRTAP1-3 | 81850 | 34 | 0 | 25 | |
| GSE47630 | KRTAP1-3 | 81850 | 8 | 1 | 31 | |
| GSE54993 | KRTAP1-3 | 81850 | 3 | 4 | 63 | |
| GSE54994 | KRTAP1-3 | 81850 | 8 | 5 | 40 | |
| GSE60625 | KRTAP1-3 | 81850 | 4 | 0 | 7 | |
| GSE74703 | KRTAP1-3 | 81850 | 1 | 1 | 34 | |
| GSE74704 | KRTAP1-3 | 81850 | 4 | 1 | 15 |
Total number of gains: 75; Total number of losses: 18; Total Number of normals: 299.
Somatic mutations of KRTAP1-3:
Generating mutation plots.
Highly correlated genes for KRTAP1-3:
Showing top 20/388 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRTAP1-3 | MAP1A | 0.76053 | 4 | 0 | 4 |
| KRTAP1-3 | KRTAP20-2 | 0.756407 | 3 | 0 | 3 |
| KRTAP1-3 | TMEM220 | 0.748776 | 3 | 0 | 3 |
| KRTAP1-3 | GFRA3 | 0.747448 | 3 | 0 | 3 |
| KRTAP1-3 | MRGPRF | 0.744668 | 3 | 0 | 3 |
| KRTAP1-3 | PDE5A | 0.729661 | 3 | 0 | 3 |
| KRTAP1-3 | WDR27 | 0.72391 | 3 | 0 | 3 |
| KRTAP1-3 | PCDH9 | 0.723902 | 3 | 0 | 3 |
| KRTAP1-3 | NAALADL1 | 0.714754 | 4 | 0 | 4 |
| KRTAP1-3 | DPP4 | 0.702588 | 3 | 0 | 3 |
| KRTAP1-3 | RUNX1T1 | 0.700228 | 3 | 0 | 3 |
| KRTAP1-3 | PDE3A | 0.700204 | 3 | 0 | 3 |
| KRTAP1-3 | EEPD1 | 0.699184 | 4 | 0 | 3 |
| KRTAP1-3 | PDE12 | 0.695229 | 4 | 0 | 4 |
| KRTAP1-3 | ASH1L | 0.684567 | 3 | 0 | 3 |
| KRTAP1-3 | GABRA5 | 0.679594 | 3 | 0 | 3 |
| KRTAP1-3 | SFTPA2 | 0.678349 | 3 | 0 | 3 |
| KRTAP1-3 | EHD2 | 0.678003 | 3 | 0 | 3 |
| KRTAP1-3 | ANK2 | 0.677517 | 3 | 0 | 3 |
| KRTAP1-3 | NMNAT2 | 0.667155 | 3 | 0 | 3 |
For details and further investigation, click here