| Full name: sucrase-isomaltase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q26.1 | ||
| Entrez ID: 6476 | HGNC ID: HGNC:10856 | Ensembl Gene: ENSG00000090402 | OMIM ID: 609845 |
| Drug and gene relationship at DGIdb | |||
Expression of SI:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SI | 6476 | 206664_at | 0.2257 | 0.4969 | |
| GSE20347 | SI | 6476 | 206664_at | 0.0112 | 0.8293 | |
| GSE23400 | SI | 6476 | 206664_at | -0.0236 | 0.1000 | |
| GSE26886 | SI | 6476 | 206664_at | 0.0054 | 0.9537 | |
| GSE29001 | SI | 6476 | 206664_at | -0.0136 | 0.9103 | |
| GSE38129 | SI | 6476 | 206664_at | -0.0120 | 0.8607 | |
| GSE45670 | SI | 6476 | 206664_at | 0.0214 | 0.7595 | |
| GSE53622 | SI | 6476 | 104829 | 0.0741 | 0.7727 | |
| GSE53624 | SI | 6476 | 104829 | 0.0929 | 0.5813 | |
| GSE63941 | SI | 6476 | 206664_at | 0.0773 | 0.5290 | |
| GSE77861 | SI | 6476 | 206664_at | 0.0535 | 0.4352 | |
| TCGA | SI | 6476 | RNAseq | 0.8630 | 0.6363 |
Upregulated datasets: 0; Downregulated datasets: 0.
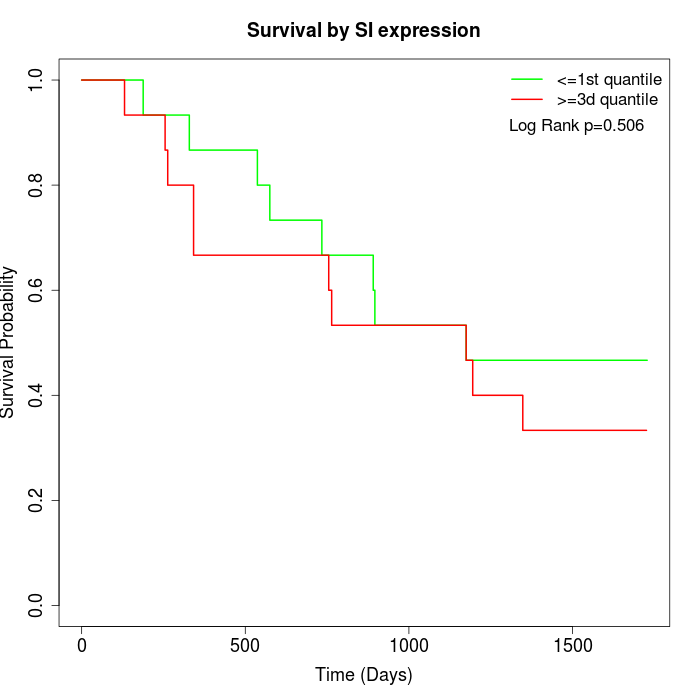
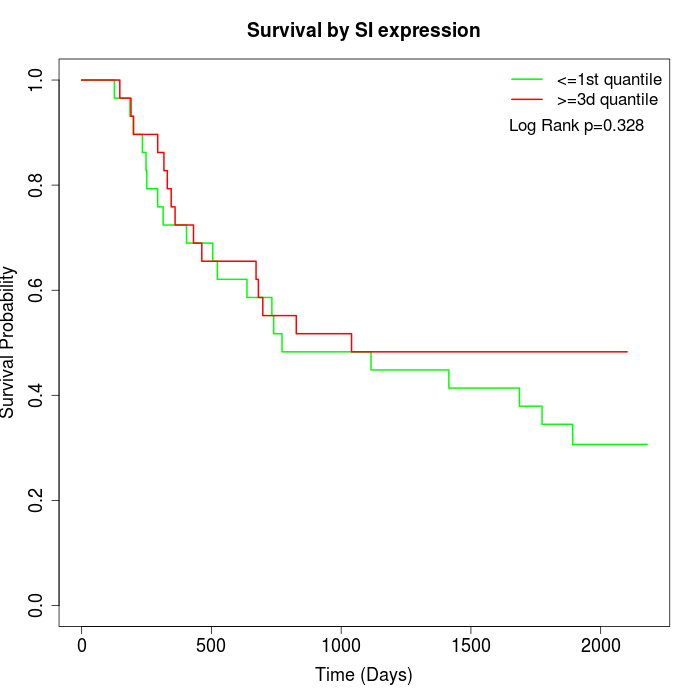
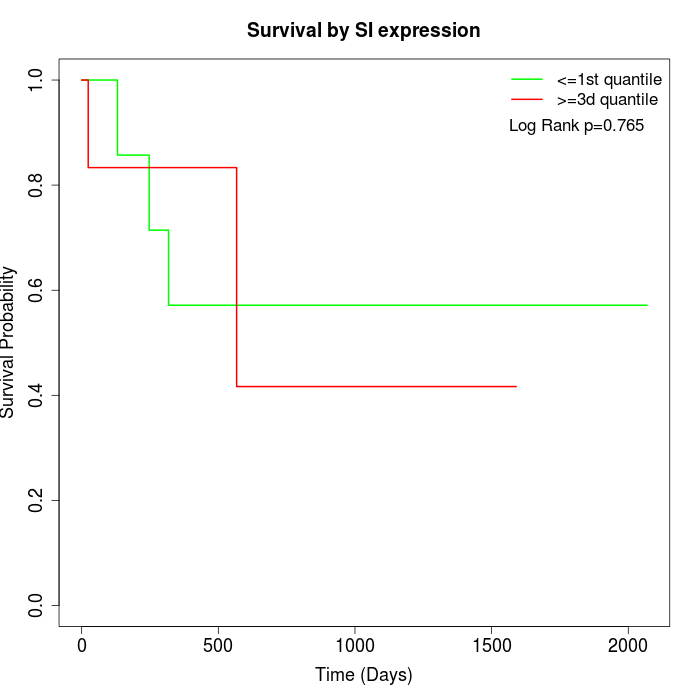
Survival by SI expression:
Note: Click image to view full size file.
Copy number change of SI:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SI | 6476 | 21 | 0 | 9 | |
| GSE20123 | SI | 6476 | 21 | 0 | 9 | |
| GSE43470 | SI | 6476 | 23 | 0 | 20 | |
| GSE46452 | SI | 6476 | 18 | 2 | 39 | |
| GSE47630 | SI | 6476 | 27 | 2 | 11 | |
| GSE54993 | SI | 6476 | 1 | 13 | 56 | |
| GSE54994 | SI | 6476 | 41 | 0 | 12 | |
| GSE60625 | SI | 6476 | 0 | 6 | 5 | |
| GSE74703 | SI | 6476 | 19 | 0 | 17 | |
| GSE74704 | SI | 6476 | 13 | 0 | 7 | |
| TCGA | SI | 6476 | 78 | 0 | 18 |
Total number of gains: 262; Total number of losses: 23; Total Number of normals: 203.
Somatic mutations of SI:
Generating mutation plots.
Highly correlated genes for SI:
Showing top 20/173 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SI | LINC00216 | 0.737907 | 3 | 0 | 3 |
| SI | KRTAP9-9 | 0.714011 | 3 | 0 | 3 |
| SI | C6orf15 | 0.707034 | 4 | 0 | 4 |
| SI | C6 | 0.671659 | 3 | 0 | 3 |
| SI | CAMK4 | 0.669143 | 3 | 0 | 3 |
| SI | CLEC4M | 0.658357 | 5 | 0 | 5 |
| SI | SCG3 | 0.652544 | 3 | 0 | 3 |
| SI | SPTA1 | 0.640297 | 4 | 0 | 4 |
| SI | GABRA5 | 0.63827 | 3 | 0 | 3 |
| SI | KLC2 | 0.633737 | 3 | 0 | 3 |
| SI | GFRA1 | 0.63297 | 3 | 0 | 3 |
| SI | NMNAT2 | 0.631332 | 3 | 0 | 3 |
| SI | GYPA | 0.629443 | 6 | 0 | 4 |
| SI | SLC9A7 | 0.627309 | 3 | 0 | 3 |
| SI | ASB4 | 0.626373 | 3 | 0 | 3 |
| SI | STRADA | 0.624622 | 3 | 0 | 3 |
| SI | ADPRH | 0.624265 | 4 | 0 | 4 |
| SI | PVALB | 0.621563 | 5 | 0 | 3 |
| SI | DGCR9 | 0.621111 | 3 | 0 | 3 |
| SI | CGA | 0.617957 | 3 | 0 | 3 |
For details and further investigation, click here