| Full name: lymphocyte transmembrane adaptor 1 | Alias Symbol: LAX|FLJ20340 | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 54900 | HGNC ID: HGNC:26005 | Ensembl Gene: ENSG00000122188 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LAX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAX1 | 54900 | 207734_at | 0.2005 | 0.8271 | |
| GSE20347 | LAX1 | 54900 | 207734_at | 0.0351 | 0.7633 | |
| GSE23400 | LAX1 | 54900 | 207734_at | 0.0219 | 0.5365 | |
| GSE26886 | LAX1 | 54900 | 207734_at | -0.1918 | 0.2074 | |
| GSE29001 | LAX1 | 54900 | 207734_at | 0.0431 | 0.7665 | |
| GSE38129 | LAX1 | 54900 | 207734_at | 0.0528 | 0.7022 | |
| GSE45670 | LAX1 | 54900 | 207734_at | 0.1246 | 0.6642 | |
| GSE53622 | LAX1 | 54900 | 72607 | 0.2858 | 0.1072 | |
| GSE53624 | LAX1 | 54900 | 72607 | 0.0461 | 0.7697 | |
| GSE63941 | LAX1 | 54900 | 207734_at | 0.1347 | 0.2644 | |
| GSE77861 | LAX1 | 54900 | 207734_at | -0.0294 | 0.8209 | |
| GSE97050 | LAX1 | 54900 | A_23_P438 | 0.6727 | 0.1909 | |
| SRP007169 | LAX1 | 54900 | RNAseq | 0.3376 | 0.6554 | |
| SRP064894 | LAX1 | 54900 | RNAseq | 0.2635 | 0.3787 | |
| SRP133303 | LAX1 | 54900 | RNAseq | 0.3683 | 0.1225 | |
| SRP159526 | LAX1 | 54900 | RNAseq | 0.3858 | 0.6374 | |
| SRP193095 | LAX1 | 54900 | RNAseq | 0.1826 | 0.4303 | |
| SRP219564 | LAX1 | 54900 | RNAseq | 0.0099 | 0.9822 | |
| TCGA | LAX1 | 54900 | RNAseq | -0.3223 | 0.2368 |
Upregulated datasets: 0; Downregulated datasets: 0.
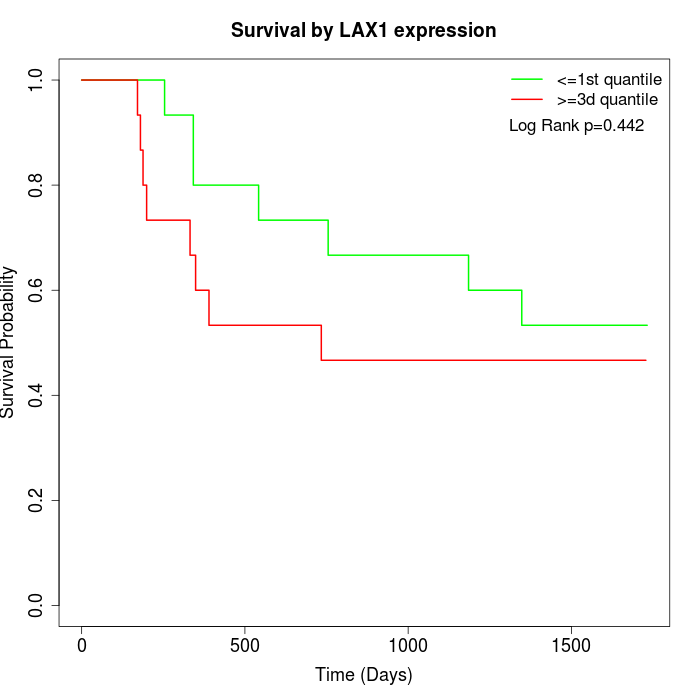
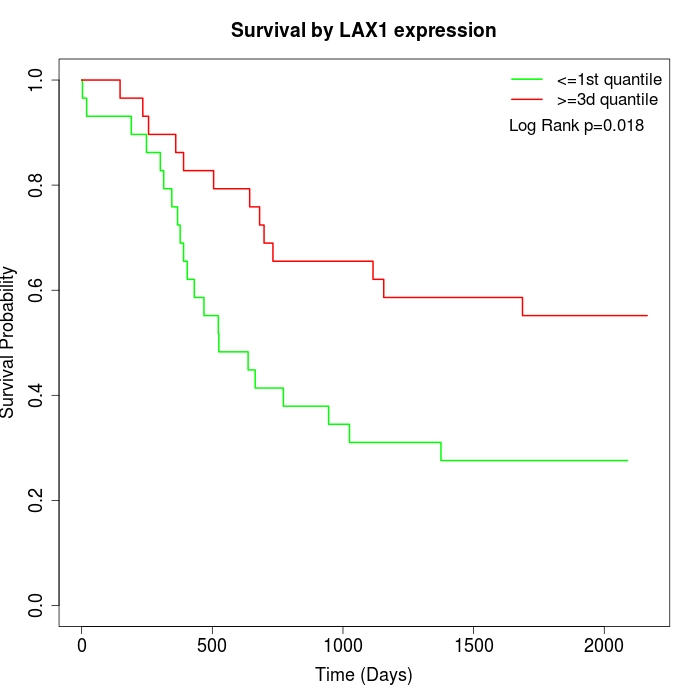
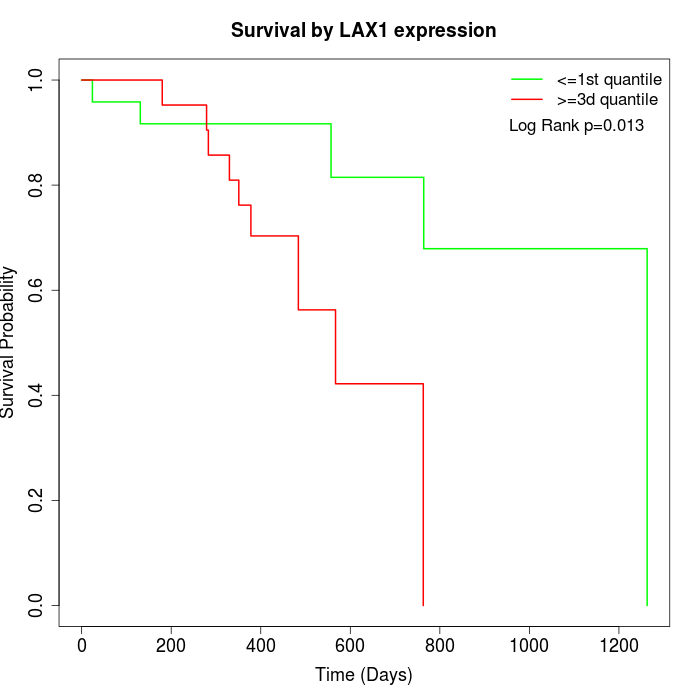
Survival by LAX1 expression:
Note: Click image to view full size file.
Copy number change of LAX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAX1 | 54900 | 11 | 0 | 19 | |
| GSE20123 | LAX1 | 54900 | 11 | 0 | 19 | |
| GSE43470 | LAX1 | 54900 | 6 | 0 | 37 | |
| GSE46452 | LAX1 | 54900 | 3 | 1 | 55 | |
| GSE47630 | LAX1 | 54900 | 14 | 0 | 26 | |
| GSE54993 | LAX1 | 54900 | 0 | 6 | 64 | |
| GSE54994 | LAX1 | 54900 | 15 | 0 | 38 | |
| GSE60625 | LAX1 | 54900 | 0 | 0 | 11 | |
| GSE74703 | LAX1 | 54900 | 6 | 0 | 30 | |
| GSE74704 | LAX1 | 54900 | 5 | 0 | 15 | |
| TCGA | LAX1 | 54900 | 43 | 4 | 49 |
Total number of gains: 114; Total number of losses: 11; Total Number of normals: 363.
Somatic mutations of LAX1:
Generating mutation plots.
Highly correlated genes for LAX1:
Showing top 20/270 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAX1 | FCRL5 | 0.77638 | 5 | 0 | 5 |
| LAX1 | MZB1 | 0.750171 | 8 | 0 | 8 |
| LAX1 | SNX20 | 0.744475 | 6 | 0 | 6 |
| LAX1 | PTPN22 | 0.7351 | 4 | 0 | 3 |
| LAX1 | POU2AF1 | 0.727745 | 4 | 0 | 4 |
| LAX1 | P2RY8 | 0.724504 | 5 | 0 | 4 |
| LAX1 | BTLA | 0.707151 | 6 | 0 | 6 |
| LAX1 | TNFRSF17 | 0.699255 | 11 | 0 | 11 |
| LAX1 | PARP11 | 0.692573 | 3 | 0 | 3 |
| LAX1 | EVI2B | 0.684301 | 10 | 0 | 10 |
| LAX1 | RASAL3 | 0.671636 | 4 | 0 | 4 |
| LAX1 | PIM2 | 0.668488 | 7 | 0 | 5 |
| LAX1 | IL16 | 0.665794 | 4 | 0 | 4 |
| LAX1 | CD79B | 0.664443 | 7 | 0 | 5 |
| LAX1 | CD180 | 0.6616 | 9 | 0 | 8 |
| LAX1 | ZBP1 | 0.655347 | 4 | 0 | 4 |
| LAX1 | IGK | 0.653176 | 6 | 0 | 6 |
| LAX1 | CD19 | 0.653129 | 8 | 0 | 6 |
| LAX1 | IKZF3 | 0.649799 | 9 | 0 | 7 |
| LAX1 | CCR5 | 0.648492 | 7 | 0 | 7 |
For details and further investigation, click here