| Full name: ladybird homeobox 1 | Alias Symbol: LBX1H|HPX6 | ||
| Type: protein-coding gene | Cytoband: 10q24.32 | ||
| Entrez ID: 10660 | HGNC ID: HGNC:16960 | Ensembl Gene: ENSG00000138136 | OMIM ID: 604255 |
| Drug and gene relationship at DGIdb | |||
Expression of LBX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LBX1 | 10660 | 208380_at | -0.0355 | 0.9245 | |
| GSE20347 | LBX1 | 10660 | 208380_at | -0.1644 | 0.0167 | |
| GSE23400 | LBX1 | 10660 | 208380_at | -0.1445 | 0.0000 | |
| GSE26886 | LBX1 | 10660 | 208380_at | 0.1447 | 0.2767 | |
| GSE29001 | LBX1 | 10660 | 208380_at | -0.1081 | 0.2030 | |
| GSE38129 | LBX1 | 10660 | 208380_at | -0.1784 | 0.0038 | |
| GSE45670 | LBX1 | 10660 | 208380_at | -0.1012 | 0.2938 | |
| GSE63941 | LBX1 | 10660 | 208380_at | 0.1535 | 0.3252 | |
| GSE77861 | LBX1 | 10660 | 208380_at | -0.1857 | 0.1186 | |
| GSE97050 | LBX1 | 10660 | A_23_P86493 | 0.0490 | 0.8293 | |
| TCGA | LBX1 | 10660 | RNAseq | 3.1309 | 0.0911 |
Upregulated datasets: 0; Downregulated datasets: 0.
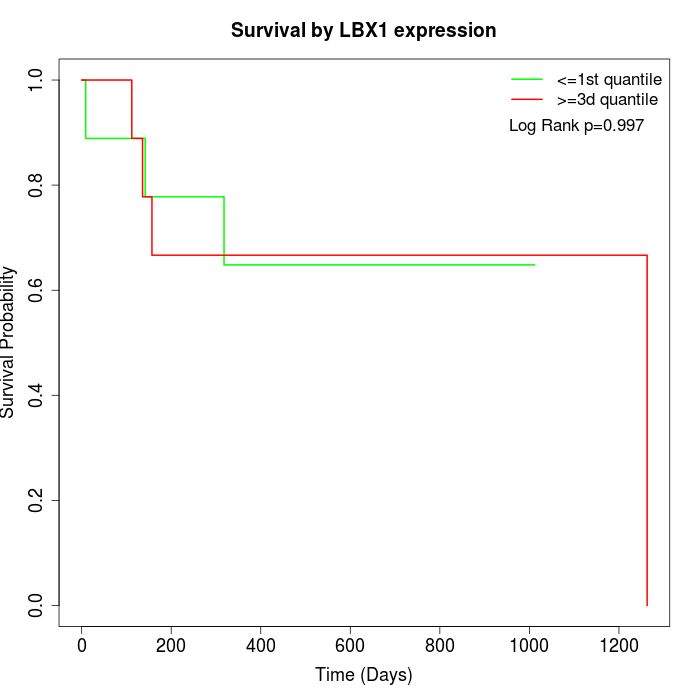
Survival by LBX1 expression:
Note: Click image to view full size file.
Copy number change of LBX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LBX1 | 10660 | 1 | 9 | 20 | |
| GSE20123 | LBX1 | 10660 | 1 | 8 | 21 | |
| GSE43470 | LBX1 | 10660 | 0 | 8 | 35 | |
| GSE46452 | LBX1 | 10660 | 0 | 11 | 48 | |
| GSE47630 | LBX1 | 10660 | 2 | 14 | 24 | |
| GSE54993 | LBX1 | 10660 | 8 | 0 | 62 | |
| GSE54994 | LBX1 | 10660 | 1 | 12 | 40 | |
| GSE60625 | LBX1 | 10660 | 0 | 0 | 11 | |
| GSE74703 | LBX1 | 10660 | 0 | 5 | 31 | |
| GSE74704 | LBX1 | 10660 | 1 | 4 | 15 | |
| TCGA | LBX1 | 10660 | 5 | 28 | 63 |
Total number of gains: 19; Total number of losses: 99; Total Number of normals: 370.
Somatic mutations of LBX1:
Generating mutation plots.
Highly correlated genes for LBX1:
Showing top 20/487 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LBX1 | MIA | 0.749211 | 3 | 0 | 3 |
| LBX1 | TMEM129 | 0.744708 | 3 | 0 | 3 |
| LBX1 | GPR31 | 0.7213 | 4 | 0 | 4 |
| LBX1 | FABP2 | 0.720088 | 3 | 0 | 3 |
| LBX1 | CNTF | 0.718448 | 3 | 0 | 3 |
| LBX1 | NDUFV3 | 0.717637 | 3 | 0 | 3 |
| LBX1 | PRPH2 | 0.709063 | 4 | 0 | 4 |
| LBX1 | TYSND1 | 0.703159 | 3 | 0 | 3 |
| LBX1 | LRRC28 | 0.702249 | 3 | 0 | 3 |
| LBX1 | SLC28A2 | 0.702146 | 3 | 0 | 3 |
| LBX1 | THRB | 0.697616 | 4 | 0 | 4 |
| LBX1 | ANO8 | 0.689483 | 3 | 0 | 3 |
| LBX1 | CCIN | 0.689144 | 5 | 0 | 4 |
| LBX1 | ADAM11 | 0.678808 | 6 | 0 | 5 |
| LBX1 | ADAMTSL5 | 0.677079 | 4 | 0 | 4 |
| LBX1 | PVR | 0.672824 | 5 | 0 | 5 |
| LBX1 | HSPB7 | 0.664868 | 3 | 0 | 3 |
| LBX1 | DDX52 | 0.663895 | 3 | 0 | 3 |
| LBX1 | INSRR | 0.662238 | 5 | 0 | 5 |
| LBX1 | USP54 | 0.66116 | 3 | 0 | 3 |
For details and further investigation, click here