| Full name: late cornified envelope 1E | Alias Symbol: LEP5 | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 353135 | HGNC ID: HGNC:29466 | Ensembl Gene: ENSG00000186226 | OMIM ID: 612607 |
| Drug and gene relationship at DGIdb | |||
Expression of LCE1E:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCE1E | 353135 | 1559226_x_at | 0.1292 | 0.5338 | |
| GSE26886 | LCE1E | 353135 | 1559226_x_at | -0.0672 | 0.5937 | |
| GSE45670 | LCE1E | 353135 | 1559226_x_at | 0.2293 | 0.0304 | |
| GSE53622 | LCE1E | 353135 | 95656 | 0.0385 | 0.5314 | |
| GSE53624 | LCE1E | 353135 | 95656 | 0.0527 | 0.7260 | |
| GSE63941 | LCE1E | 353135 | 1559226_x_at | 0.1125 | 0.4898 | |
| GSE77861 | LCE1E | 353135 | 1559226_x_at | 0.0094 | 0.9629 | |
| GSE97050 | LCE1E | 353135 | A_33_P3356811 | -0.0228 | 0.9340 | |
| TCGA | LCE1E | 353135 | RNAseq | 3.0261 | 0.0050 |
Upregulated datasets: 1; Downregulated datasets: 0.
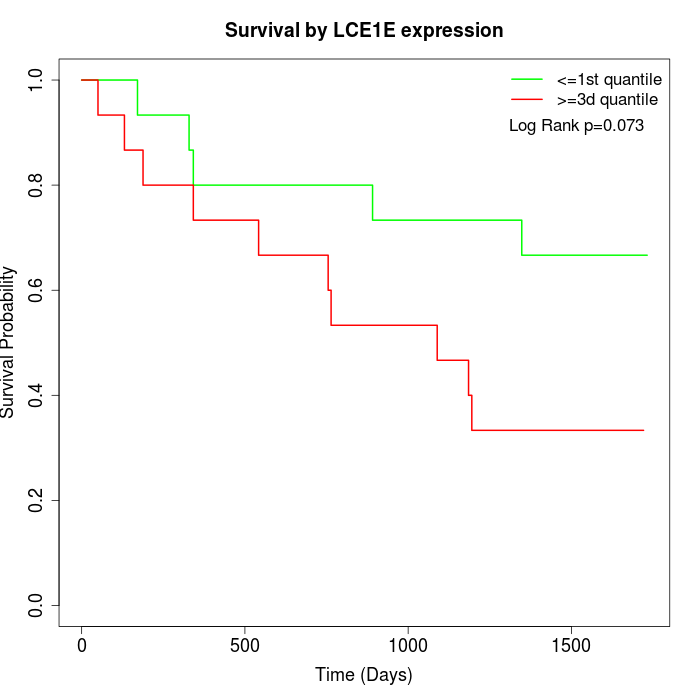
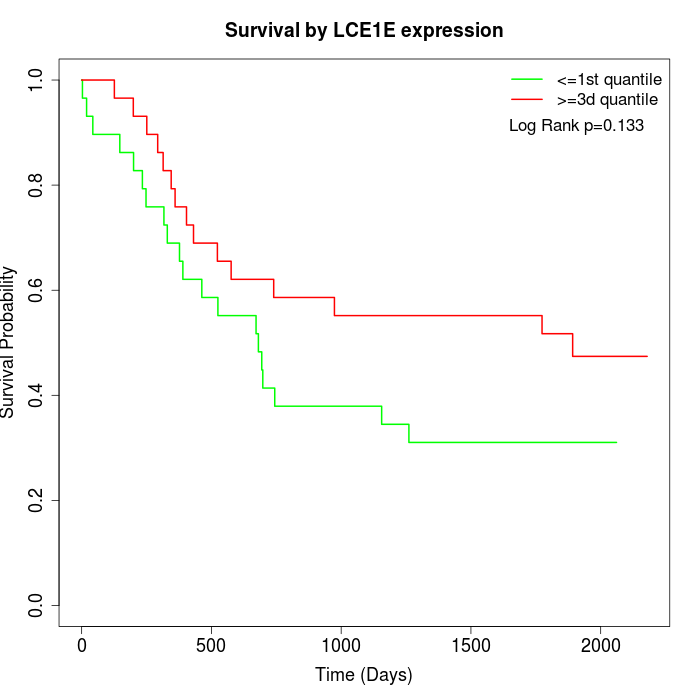
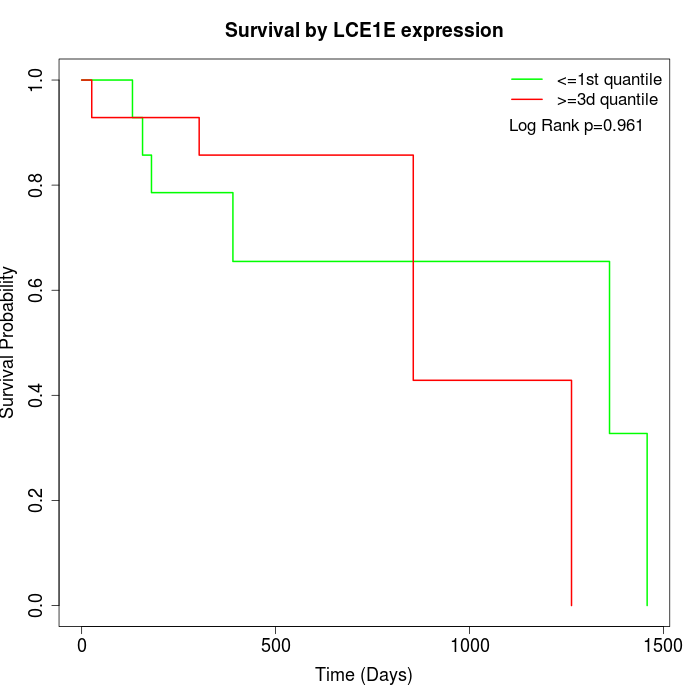
Survival by LCE1E expression:
Note: Click image to view full size file.
Copy number change of LCE1E:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCE1E | 353135 | 15 | 0 | 15 | |
| GSE20123 | LCE1E | 353135 | 15 | 0 | 15 | |
| GSE43470 | LCE1E | 353135 | 6 | 2 | 35 | |
| GSE46452 | LCE1E | 353135 | 3 | 1 | 55 | |
| GSE47630 | LCE1E | 353135 | 15 | 0 | 25 | |
| GSE54993 | LCE1E | 353135 | 0 | 4 | 66 | |
| GSE54994 | LCE1E | 353135 | 15 | 0 | 38 | |
| GSE60625 | LCE1E | 353135 | 0 | 0 | 11 | |
| GSE74703 | LCE1E | 353135 | 6 | 1 | 29 | |
| GSE74704 | LCE1E | 353135 | 7 | 0 | 13 | |
| TCGA | LCE1E | 353135 | 39 | 2 | 55 |
Total number of gains: 121; Total number of losses: 10; Total Number of normals: 357.
Somatic mutations of LCE1E:
Generating mutation plots.
Highly correlated genes for LCE1E:
Showing top 20/101 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCE1E | LYSMD1 | 0.737174 | 3 | 0 | 3 |
| LCE1E | DCHS2 | 0.729579 | 3 | 0 | 3 |
| LCE1E | ZNF461 | 0.720553 | 3 | 0 | 3 |
| LCE1E | SNAI3 | 0.702322 | 3 | 0 | 3 |
| LCE1E | SPRR4 | 0.6854 | 3 | 0 | 3 |
| LCE1E | MAPK12 | 0.684278 | 3 | 0 | 3 |
| LCE1E | KCNQ3 | 0.680595 | 3 | 0 | 3 |
| LCE1E | LRRD1 | 0.680152 | 4 | 0 | 3 |
| LCE1E | RAB11FIP3 | 0.679129 | 3 | 0 | 3 |
| LCE1E | TFRC | 0.677071 | 3 | 0 | 3 |
| LCE1E | C20orf96 | 0.673446 | 3 | 0 | 3 |
| LCE1E | DNAH1 | 0.671206 | 4 | 0 | 3 |
| LCE1E | GRIA4 | 0.665002 | 3 | 0 | 3 |
| LCE1E | CNTNAP1 | 0.659976 | 4 | 0 | 3 |
| LCE1E | APOBEC2 | 0.659491 | 4 | 0 | 3 |
| LCE1E | LONRF3 | 0.656461 | 4 | 0 | 3 |
| LCE1E | PCDH12 | 0.648631 | 3 | 0 | 3 |
| LCE1E | CCM2L | 0.648556 | 4 | 0 | 3 |
| LCE1E | CSF3R | 0.643041 | 6 | 0 | 4 |
| LCE1E | ZC3H18 | 0.638288 | 4 | 0 | 3 |
For details and further investigation, click here