| Full name: lysocardiolipin acyltransferase 1 | Alias Symbol: FLJ37965|ALCAT1|AGPAT8 | ||
| Type: protein-coding gene | Cytoband: 2p23.1 | ||
| Entrez ID: 253558 | HGNC ID: HGNC:26756 | Ensembl Gene: ENSG00000172954 | OMIM ID: 614241 |
| Drug and gene relationship at DGIdb | |||
Expression of LCLAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCLAT1 | 253558 | 226996_at | 0.4279 | 0.4280 | |
| GSE26886 | LCLAT1 | 253558 | 226996_at | 0.9921 | 0.0003 | |
| GSE45670 | LCLAT1 | 253558 | 226996_at | 0.6913 | 0.0001 | |
| GSE53622 | LCLAT1 | 253558 | 129220 | 0.7118 | 0.0000 | |
| GSE53624 | LCLAT1 | 253558 | 87955 | 0.6353 | 0.0000 | |
| GSE63941 | LCLAT1 | 253558 | 226996_at | 0.1969 | 0.6165 | |
| GSE77861 | LCLAT1 | 253558 | 226996_at | 1.0114 | 0.0017 | |
| GSE97050 | LCLAT1 | 253558 | A_33_P3349693 | 0.4932 | 0.3085 | |
| SRP007169 | LCLAT1 | 253558 | RNAseq | 1.3887 | 0.0003 | |
| SRP008496 | LCLAT1 | 253558 | RNAseq | 1.6537 | 0.0000 | |
| SRP064894 | LCLAT1 | 253558 | RNAseq | 0.6886 | 0.0000 | |
| SRP133303 | LCLAT1 | 253558 | RNAseq | 1.1648 | 0.0000 | |
| SRP159526 | LCLAT1 | 253558 | RNAseq | 1.1456 | 0.0000 | |
| SRP193095 | LCLAT1 | 253558 | RNAseq | 0.6573 | 0.0000 | |
| SRP219564 | LCLAT1 | 253558 | RNAseq | 0.2088 | 0.6100 | |
| TCGA | LCLAT1 | 253558 | RNAseq | 0.1963 | 0.0005 |
Upregulated datasets: 5; Downregulated datasets: 0.
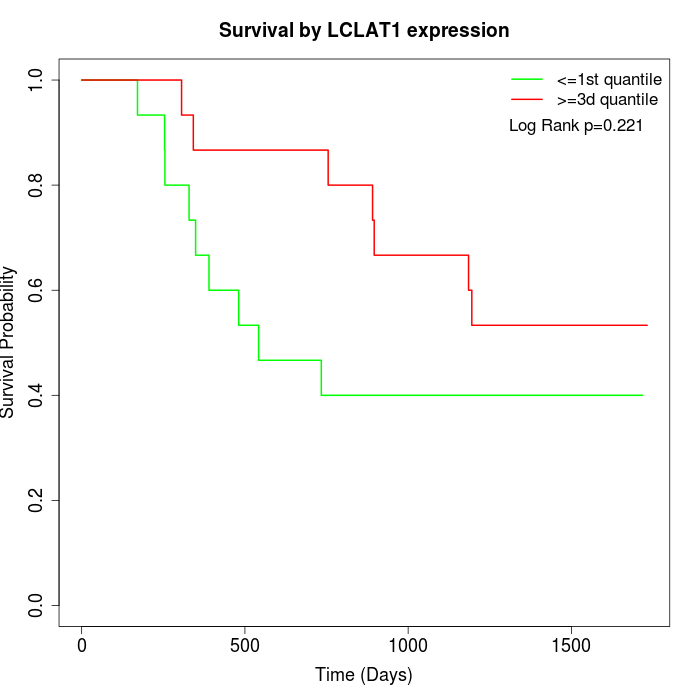
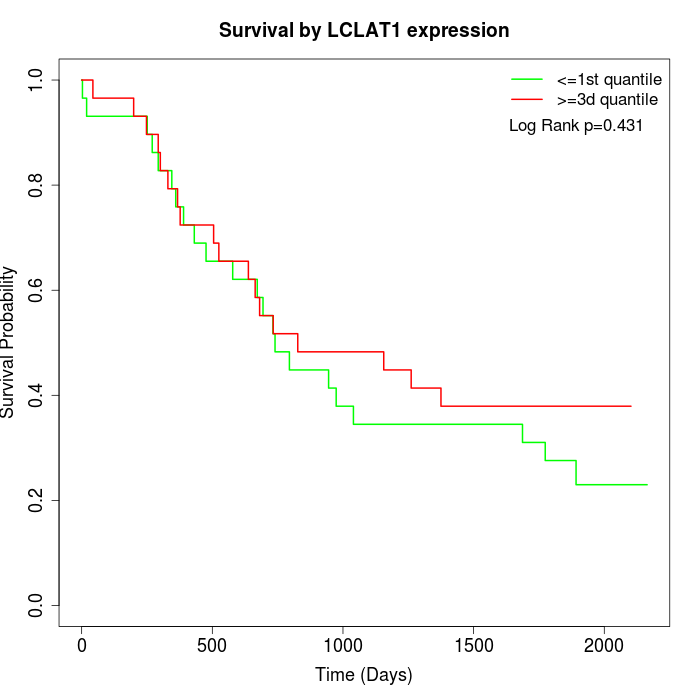
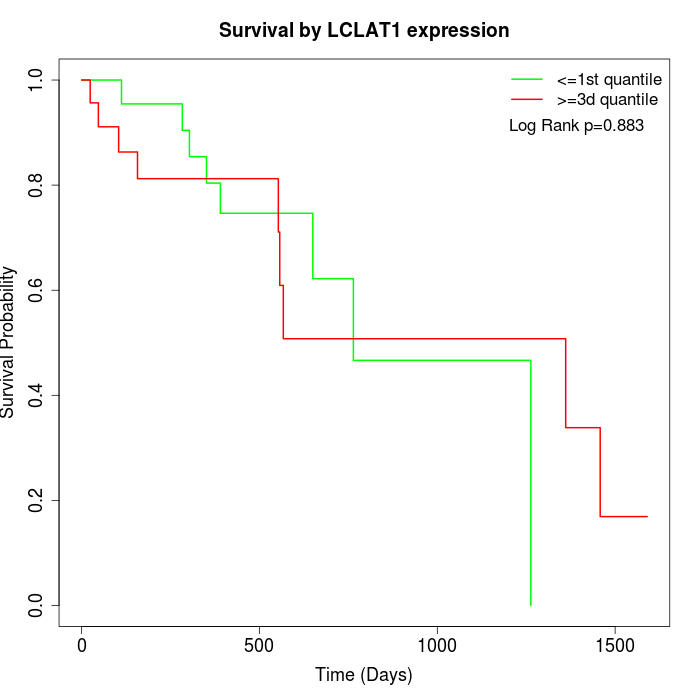
Survival by LCLAT1 expression:
Note: Click image to view full size file.
Copy number change of LCLAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCLAT1 | 253558 | 11 | 1 | 18 | |
| GSE20123 | LCLAT1 | 253558 | 9 | 1 | 20 | |
| GSE43470 | LCLAT1 | 253558 | 2 | 1 | 40 | |
| GSE46452 | LCLAT1 | 253558 | 2 | 4 | 53 | |
| GSE47630 | LCLAT1 | 253558 | 8 | 0 | 32 | |
| GSE54993 | LCLAT1 | 253558 | 0 | 6 | 64 | |
| GSE54994 | LCLAT1 | 253558 | 10 | 0 | 43 | |
| GSE60625 | LCLAT1 | 253558 | 0 | 3 | 8 | |
| GSE74703 | LCLAT1 | 253558 | 2 | 0 | 34 | |
| GSE74704 | LCLAT1 | 253558 | 9 | 1 | 10 | |
| TCGA | LCLAT1 | 253558 | 36 | 2 | 58 |
Total number of gains: 89; Total number of losses: 19; Total Number of normals: 380.
Somatic mutations of LCLAT1:
Generating mutation plots.
Highly correlated genes for LCLAT1:
Showing top 20/1665 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCLAT1 | CUX1 | 0.785178 | 6 | 0 | 6 |
| LCLAT1 | A4GALT | 0.762099 | 3 | 0 | 3 |
| LCLAT1 | CD44 | 0.757691 | 3 | 0 | 3 |
| LCLAT1 | PDPR | 0.754094 | 5 | 0 | 5 |
| LCLAT1 | C1orf226 | 0.74184 | 4 | 0 | 3 |
| LCLAT1 | PARP2 | 0.740233 | 3 | 0 | 3 |
| LCLAT1 | RAB34 | 0.738199 | 3 | 0 | 3 |
| LCLAT1 | MFHAS1 | 0.736887 | 7 | 0 | 7 |
| LCLAT1 | STK3 | 0.735719 | 5 | 0 | 5 |
| LCLAT1 | MORC2 | 0.734068 | 5 | 0 | 4 |
| LCLAT1 | ZBED1 | 0.725895 | 3 | 0 | 3 |
| LCLAT1 | ZFHX3 | 0.723025 | 4 | 0 | 3 |
| LCLAT1 | ASB7 | 0.722157 | 4 | 0 | 3 |
| LCLAT1 | TSC22D1 | 0.721282 | 3 | 0 | 3 |
| LCLAT1 | SUPT7L | 0.719668 | 6 | 0 | 6 |
| LCLAT1 | LPCAT1 | 0.719346 | 6 | 0 | 6 |
| LCLAT1 | ATP6V1C1 | 0.717935 | 5 | 0 | 5 |
| LCLAT1 | FADS1 | 0.717362 | 3 | 0 | 3 |
| LCLAT1 | EPPK1 | 0.716189 | 5 | 0 | 5 |
| LCLAT1 | NF1 | 0.714752 | 3 | 0 | 3 |
For details and further investigation, click here