| Full name: lactate dehydrogenase A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11p15.1 | ||
| Entrez ID: 3939 | HGNC ID: HGNC:6535 | Ensembl Gene: ENSG00000134333 | OMIM ID: 150000 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
LDHA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of LDHA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LDHA | 3939 | 200650_s_at | 0.0815 | 0.8663 | |
| GSE20347 | LDHA | 3939 | 200650_s_at | -0.1284 | 0.4437 | |
| GSE23400 | LDHA | 3939 | 200650_s_at | 0.3212 | 0.0000 | |
| GSE26886 | LDHA | 3939 | 200650_s_at | -0.2226 | 0.1357 | |
| GSE29001 | LDHA | 3939 | 200650_s_at | -0.1106 | 0.7168 | |
| GSE38129 | LDHA | 3939 | 200650_s_at | 0.1241 | 0.3399 | |
| GSE45670 | LDHA | 3939 | 200650_s_at | 0.3187 | 0.0191 | |
| GSE53622 | LDHA | 3939 | 64568 | 0.5840 | 0.0000 | |
| GSE53624 | LDHA | 3939 | 64568 | 0.6363 | 0.0000 | |
| GSE63941 | LDHA | 3939 | 200650_s_at | -0.2942 | 0.6265 | |
| GSE77861 | LDHA | 3939 | 200650_s_at | 0.1851 | 0.2622 | |
| GSE97050 | LDHA | 3939 | A_23_P47565 | 0.6577 | 0.0950 | |
| SRP007169 | LDHA | 3939 | RNAseq | -0.1630 | 0.5946 | |
| SRP008496 | LDHA | 3939 | RNAseq | -0.0124 | 0.9461 | |
| SRP064894 | LDHA | 3939 | RNAseq | 0.3445 | 0.0865 | |
| SRP133303 | LDHA | 3939 | RNAseq | 1.0066 | 0.0003 | |
| SRP159526 | LDHA | 3939 | RNAseq | -0.1676 | 0.7007 | |
| SRP193095 | LDHA | 3939 | RNAseq | 0.1604 | 0.4190 | |
| SRP219564 | LDHA | 3939 | RNAseq | 0.1800 | 0.3330 | |
| TCGA | LDHA | 3939 | RNAseq | 0.2891 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
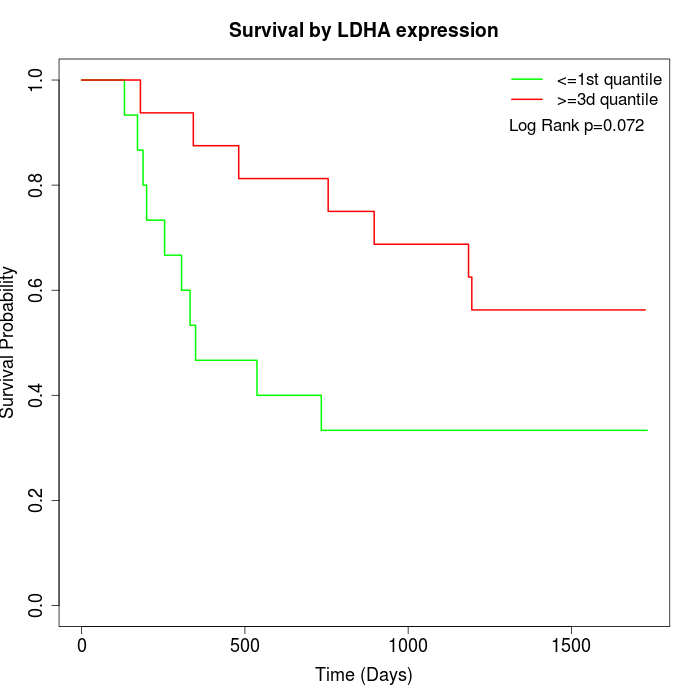
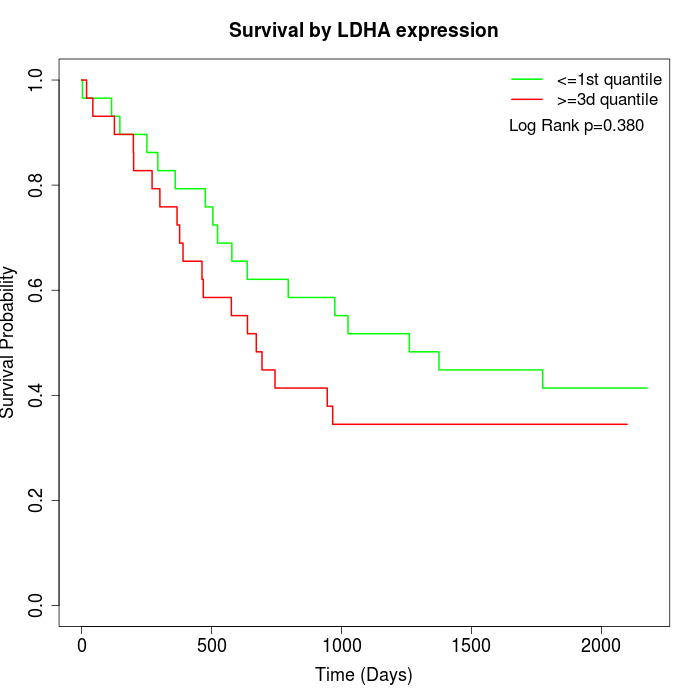
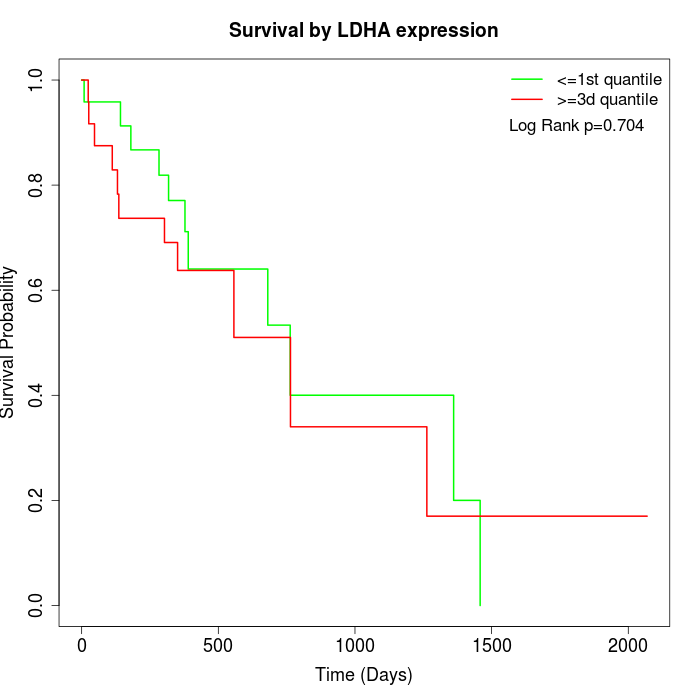
Survival by LDHA expression:
Note: Click image to view full size file.
Copy number change of LDHA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LDHA | 3939 | 0 | 10 | 20 | |
| GSE20123 | LDHA | 3939 | 0 | 10 | 20 | |
| GSE43470 | LDHA | 3939 | 1 | 5 | 37 | |
| GSE46452 | LDHA | 3939 | 7 | 5 | 47 | |
| GSE47630 | LDHA | 3939 | 3 | 10 | 27 | |
| GSE54993 | LDHA | 3939 | 3 | 0 | 67 | |
| GSE54994 | LDHA | 3939 | 2 | 13 | 38 | |
| GSE60625 | LDHA | 3939 | 0 | 0 | 11 | |
| GSE74703 | LDHA | 3939 | 1 | 3 | 32 | |
| GSE74704 | LDHA | 3939 | 0 | 9 | 11 | |
| TCGA | LDHA | 3939 | 11 | 29 | 56 |
Total number of gains: 28; Total number of losses: 94; Total Number of normals: 366.
Somatic mutations of LDHA:
Generating mutation plots.
Highly correlated genes for LDHA:
Showing top 20/531 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LDHA | GTF3C6 | 0.731022 | 3 | 0 | 3 |
| LDHA | PRKAA1 | 0.698609 | 3 | 0 | 3 |
| LDHA | RRAS2 | 0.698385 | 4 | 0 | 4 |
| LDHA | STYX | 0.694592 | 3 | 0 | 3 |
| LDHA | SRD5A1 | 0.690606 | 3 | 0 | 3 |
| LDHA | REEP3 | 0.689279 | 3 | 0 | 3 |
| LDHA | NEK6 | 0.676798 | 4 | 0 | 3 |
| LDHA | CRIPT | 0.670509 | 4 | 0 | 3 |
| LDHA | SEC61G | 0.669339 | 7 | 0 | 7 |
| LDHA | SERPINB9 | 0.667072 | 4 | 0 | 3 |
| LDHA | TMEM167A | 0.663653 | 5 | 0 | 4 |
| LDHA | CDK18 | 0.663206 | 3 | 0 | 3 |
| LDHA | MALT1 | 0.660908 | 6 | 0 | 5 |
| LDHA | PGK1 | 0.658385 | 12 | 0 | 12 |
| LDHA | AGTRAP | 0.657495 | 3 | 0 | 3 |
| LDHA | SPON2 | 0.65593 | 3 | 0 | 3 |
| LDHA | RRAGC | 0.653822 | 3 | 0 | 3 |
| LDHA | TRAPPC6B | 0.652357 | 3 | 0 | 3 |
| LDHA | EBNA1BP2 | 0.649974 | 4 | 0 | 3 |
| LDHA | ADAMTS14 | 0.649849 | 3 | 0 | 3 |
For details and further investigation, click here