| Full name: long intergenic non-protein coding RNA 700 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 10p15.3 | ||
| Entrez ID: 282980 | HGNC ID: HGNC:27422 | Ensembl Gene: ENSG00000234962 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00700:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00700 | 282980 | 1564209_at | 0.0083 | 0.9830 | |
| GSE26886 | LINC00700 | 282980 | 1564209_at | 0.2108 | 0.0067 | |
| GSE45670 | LINC00700 | 282980 | 1564209_at | 0.0250 | 0.7980 | |
| GSE53622 | LINC00700 | 282980 | 58001 | -1.0321 | 0.0000 | |
| GSE53624 | LINC00700 | 282980 | 58001 | -2.5265 | 0.0000 | |
| GSE63941 | LINC00700 | 282980 | 1564209_at | 0.0919 | 0.5293 | |
| GSE77861 | LINC00700 | 282980 | 1564209_at | -0.0712 | 0.5288 | |
| SRP133303 | LINC00700 | 282980 | RNAseq | -0.2521 | 0.4070 |
Upregulated datasets: 0; Downregulated datasets: 2.
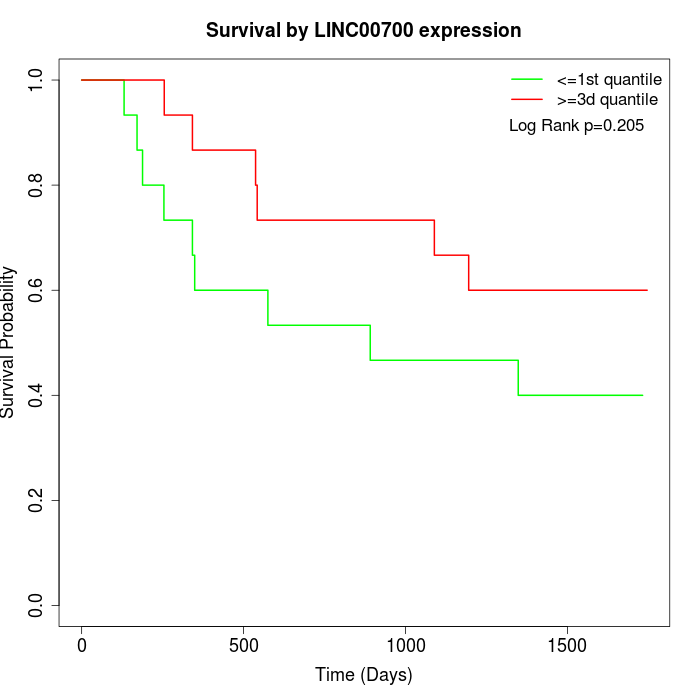
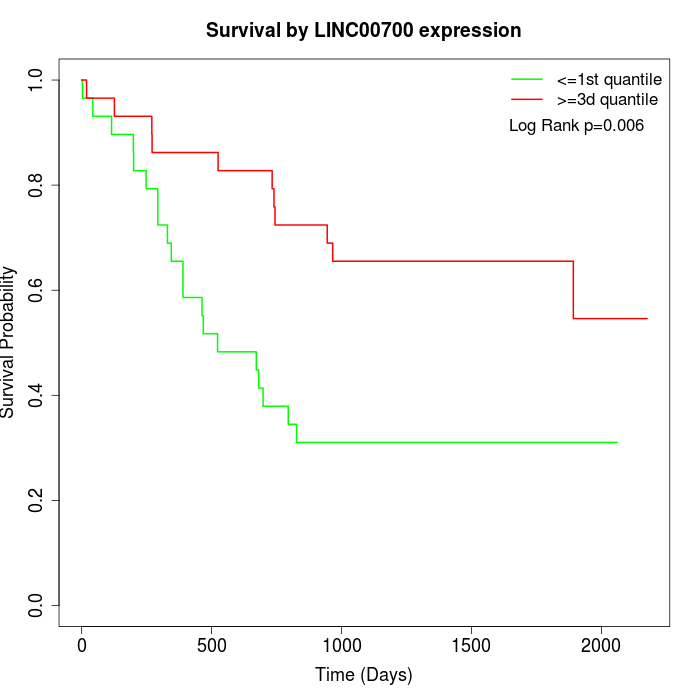
Survival by LINC00700 expression:
Note: Click image to view full size file.
Copy number change of LINC00700:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00700 | 282980 | 4 | 9 | 17 | |
| GSE20123 | LINC00700 | 282980 | 4 | 9 | 17 | |
| GSE43470 | LINC00700 | 282980 | 3 | 8 | 32 | |
| GSE46452 | LINC00700 | 282980 | 2 | 14 | 43 | |
| GSE47630 | LINC00700 | 282980 | 5 | 15 | 20 | |
| GSE54993 | LINC00700 | 282980 | 11 | 0 | 59 | |
| GSE54994 | LINC00700 | 282980 | 4 | 10 | 39 | |
| GSE60625 | LINC00700 | 282980 | 0 | 0 | 11 | |
| GSE74703 | LINC00700 | 282980 | 2 | 5 | 29 | |
| GSE74704 | LINC00700 | 282980 | 1 | 8 | 11 | |
| TCGA | LINC00700 | 282980 | 18 | 26 | 52 |
Total number of gains: 54; Total number of losses: 104; Total Number of normals: 330.
Somatic mutations of LINC00700:
Generating mutation plots.
Highly correlated genes for LINC00700:
Showing top 20/73 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00700 | CCDC172 | 0.760465 | 3 | 0 | 3 |
| LINC00700 | DCD | 0.706554 | 4 | 0 | 4 |
| LINC00700 | PXMP2 | 0.698028 | 3 | 0 | 3 |
| LINC00700 | CRX | 0.69776 | 3 | 0 | 3 |
| LINC00700 | ARID5A | 0.695598 | 4 | 0 | 4 |
| LINC00700 | C19orf57 | 0.685635 | 4 | 0 | 3 |
| LINC00700 | SPHKAP | 0.685228 | 3 | 0 | 3 |
| LINC00700 | PHF12 | 0.667789 | 3 | 0 | 3 |
| LINC00700 | SPACA4 | 0.665394 | 3 | 0 | 3 |
| LINC00700 | HYAL1 | 0.659507 | 3 | 0 | 3 |
| LINC00700 | CEACAM3 | 0.657124 | 5 | 0 | 3 |
| LINC00700 | ZCCHC13 | 0.655849 | 3 | 0 | 3 |
| LINC00700 | SFXN3 | 0.65445 | 3 | 0 | 3 |
| LINC00700 | MPO | 0.642822 | 4 | 0 | 4 |
| LINC00700 | C14orf180 | 0.640212 | 3 | 0 | 3 |
| LINC00700 | P2RX3 | 0.638506 | 4 | 0 | 3 |
| LINC00700 | ATP2B3 | 0.638295 | 3 | 0 | 3 |
| LINC00700 | GHRHR | 0.628184 | 3 | 0 | 3 |
| LINC00700 | ZDHHC22 | 0.627204 | 3 | 0 | 3 |
| LINC00700 | ZFR2 | 0.625971 | 4 | 0 | 3 |
For details and further investigation, click here