| Full name: lysophosphatidic acid receptor 2 | Alias Symbol: EDG-4|LPA2 | ||
| Type: protein-coding gene | Cytoband: 19p12 | ||
| Entrez ID: 9170 | HGNC ID: HGNC:3168 | Ensembl Gene: ENSG00000064547 | OMIM ID: 605110 |
| Drug and gene relationship at DGIdb | |||
LPAR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04072 | Phospholipase D signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway |
Expression of LPAR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LPAR2 | 9170 | 206723_s_at | 0.3496 | 0.3544 | |
| GSE20347 | LPAR2 | 9170 | 206723_s_at | 0.0704 | 0.5633 | |
| GSE23400 | LPAR2 | 9170 | 206723_s_at | 0.1404 | 0.0144 | |
| GSE26886 | LPAR2 | 9170 | 206723_s_at | 0.7038 | 0.0022 | |
| GSE29001 | LPAR2 | 9170 | 206723_s_at | 0.4034 | 0.1051 | |
| GSE38129 | LPAR2 | 9170 | 206723_s_at | 0.4554 | 0.0013 | |
| GSE45670 | LPAR2 | 9170 | 206723_s_at | 0.4580 | 0.0006 | |
| GSE53622 | LPAR2 | 9170 | 76511 | 0.3513 | 0.0002 | |
| GSE53624 | LPAR2 | 9170 | 76511 | 0.2337 | 0.0004 | |
| GSE63941 | LPAR2 | 9170 | 206723_s_at | 0.8935 | 0.0614 | |
| GSE77861 | LPAR2 | 9170 | 206723_s_at | 0.1699 | 0.1471 | |
| GSE97050 | LPAR2 | 9170 | A_23_P101829 | 0.7361 | 0.2010 | |
| SRP007169 | LPAR2 | 9170 | RNAseq | -0.7878 | 0.0839 | |
| SRP008496 | LPAR2 | 9170 | RNAseq | -0.3987 | 0.1471 | |
| SRP064894 | LPAR2 | 9170 | RNAseq | 1.2387 | 0.0001 | |
| SRP133303 | LPAR2 | 9170 | RNAseq | -0.0134 | 0.9471 | |
| SRP159526 | LPAR2 | 9170 | RNAseq | 0.2564 | 0.4451 | |
| SRP193095 | LPAR2 | 9170 | RNAseq | 0.0453 | 0.7896 | |
| SRP219564 | LPAR2 | 9170 | RNAseq | 0.7954 | 0.1027 | |
| TCGA | LPAR2 | 9170 | RNAseq | 0.0662 | 0.4428 |
Upregulated datasets: 1; Downregulated datasets: 0.
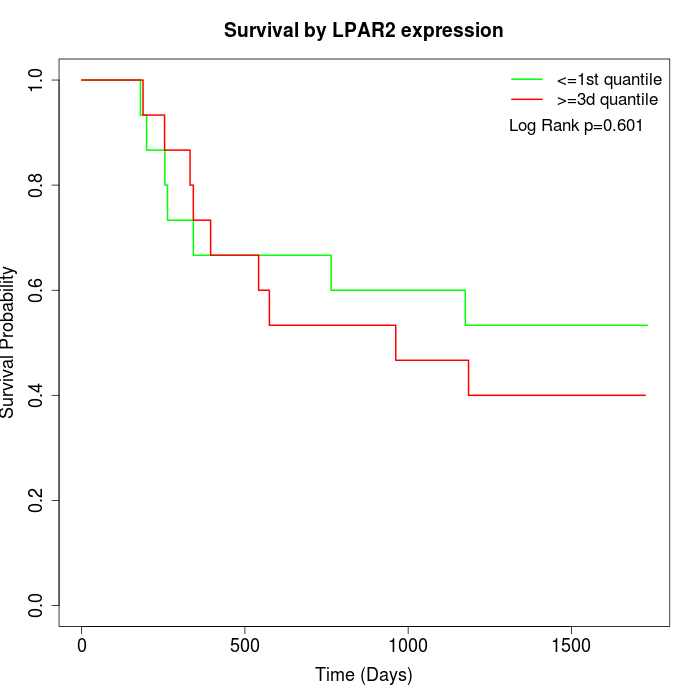
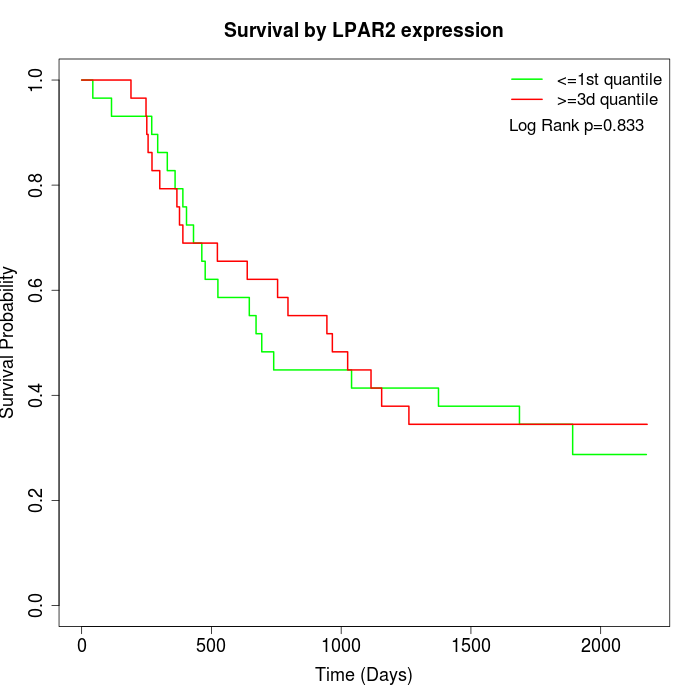
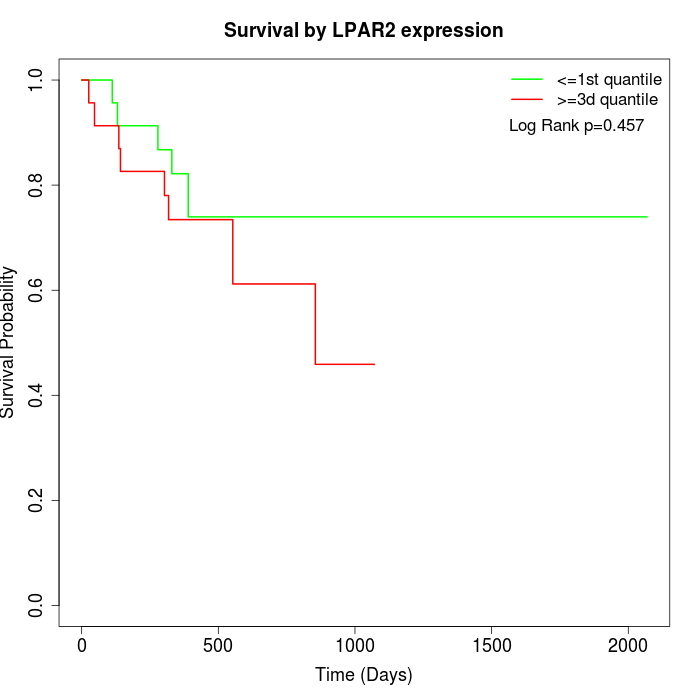
Survival by LPAR2 expression:
Note: Click image to view full size file.
Copy number change of LPAR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LPAR2 | 9170 | 4 | 5 | 21 | |
| GSE20123 | LPAR2 | 9170 | 3 | 3 | 24 | |
| GSE43470 | LPAR2 | 9170 | 2 | 5 | 36 | |
| GSE46452 | LPAR2 | 9170 | 47 | 1 | 11 | |
| GSE47630 | LPAR2 | 9170 | 5 | 8 | 27 | |
| GSE54993 | LPAR2 | 9170 | 16 | 4 | 50 | |
| GSE54994 | LPAR2 | 9170 | 6 | 13 | 34 | |
| GSE60625 | LPAR2 | 9170 | 9 | 0 | 2 | |
| GSE74703 | LPAR2 | 9170 | 2 | 3 | 31 | |
| GSE74704 | LPAR2 | 9170 | 0 | 3 | 17 | |
| TCGA | LPAR2 | 9170 | 16 | 10 | 70 |
Total number of gains: 110; Total number of losses: 55; Total Number of normals: 323.
Somatic mutations of LPAR2:
Generating mutation plots.
Highly correlated genes for LPAR2:
Showing top 20/418 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LPAR2 | ZFYVE27 | 0.749942 | 3 | 0 | 3 |
| LPAR2 | COX17 | 0.670109 | 3 | 0 | 3 |
| LPAR2 | GNPNAT1 | 0.664611 | 3 | 0 | 3 |
| LPAR2 | WIZ | 0.660098 | 5 | 0 | 5 |
| LPAR2 | KDF1 | 0.65672 | 3 | 0 | 3 |
| LPAR2 | IRX2 | 0.654419 | 5 | 0 | 4 |
| LPAR2 | POLR1A | 0.653818 | 3 | 0 | 3 |
| LPAR2 | F11R | 0.649255 | 8 | 0 | 7 |
| LPAR2 | CHCHD6 | 0.642781 | 3 | 0 | 3 |
| LPAR2 | YDJC | 0.638207 | 5 | 0 | 3 |
| LPAR2 | MTG1 | 0.637732 | 4 | 0 | 4 |
| LPAR2 | TP53I13 | 0.636297 | 3 | 0 | 3 |
| LPAR2 | C12orf73 | 0.635395 | 3 | 0 | 3 |
| LPAR2 | ELAC2 | 0.634944 | 4 | 0 | 3 |
| LPAR2 | PHF12 | 0.633847 | 3 | 0 | 3 |
| LPAR2 | ABRACL | 0.633354 | 4 | 0 | 3 |
| LPAR2 | ARHGAP23 | 0.62907 | 3 | 0 | 3 |
| LPAR2 | LIG1 | 0.628256 | 9 | 0 | 7 |
| LPAR2 | SEPHS1 | 0.626158 | 3 | 0 | 3 |
| LPAR2 | SLC25A28 | 0.625485 | 4 | 0 | 4 |
For details and further investigation, click here