| Full name: translocation associated membrane protein 2 | Alias Symbol: KIAA0057 | ||
| Type: protein-coding gene | Cytoband: 6p12.2 | ||
| Entrez ID: 9697 | HGNC ID: HGNC:16855 | Ensembl Gene: ENSG00000065308 | OMIM ID: 608485 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRAM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRAM2 | 9697 | 202369_s_at | 1.0257 | 0.0877 | |
| GSE20347 | TRAM2 | 9697 | 202369_s_at | 1.4646 | 0.0000 | |
| GSE23400 | TRAM2 | 9697 | 202369_s_at | 0.8474 | 0.0000 | |
| GSE26886 | TRAM2 | 9697 | 202369_s_at | 2.0415 | 0.0000 | |
| GSE29001 | TRAM2 | 9697 | 202369_s_at | 1.3927 | 0.0000 | |
| GSE38129 | TRAM2 | 9697 | 202369_s_at | 1.1635 | 0.0000 | |
| GSE45670 | TRAM2 | 9697 | 202369_s_at | 0.6929 | 0.0422 | |
| GSE53622 | TRAM2 | 9697 | 126510 | 1.2644 | 0.0000 | |
| GSE53624 | TRAM2 | 9697 | 126510 | 1.6112 | 0.0000 | |
| GSE63941 | TRAM2 | 9697 | 202369_s_at | -1.5474 | 0.0190 | |
| GSE77861 | TRAM2 | 9697 | 202369_s_at | 1.6394 | 0.0198 | |
| GSE97050 | TRAM2 | 9697 | A_24_P12413 | 0.3749 | 0.1506 | |
| SRP007169 | TRAM2 | 9697 | RNAseq | 3.9582 | 0.0000 | |
| SRP008496 | TRAM2 | 9697 | RNAseq | 3.4571 | 0.0000 | |
| SRP064894 | TRAM2 | 9697 | RNAseq | 1.3775 | 0.0000 | |
| SRP133303 | TRAM2 | 9697 | RNAseq | 1.1331 | 0.0000 | |
| SRP159526 | TRAM2 | 9697 | RNAseq | 1.5793 | 0.0000 | |
| SRP193095 | TRAM2 | 9697 | RNAseq | 1.3962 | 0.0000 | |
| SRP219564 | TRAM2 | 9697 | RNAseq | 1.2050 | 0.0017 | |
| TCGA | TRAM2 | 9697 | RNAseq | 0.0617 | 0.3847 |
Upregulated datasets: 14; Downregulated datasets: 1.
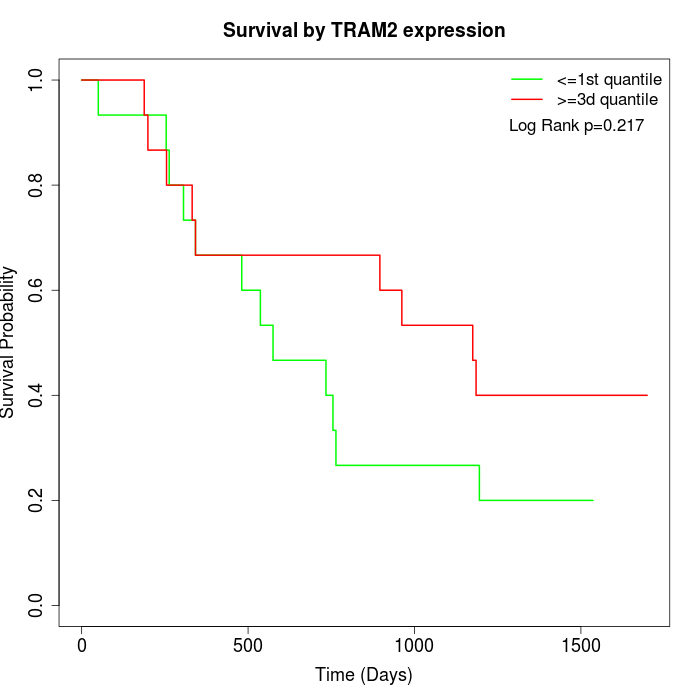
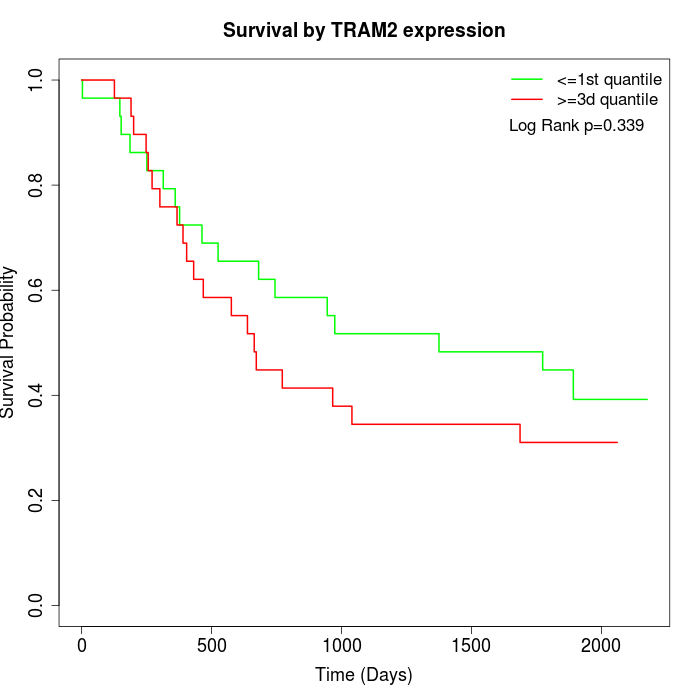
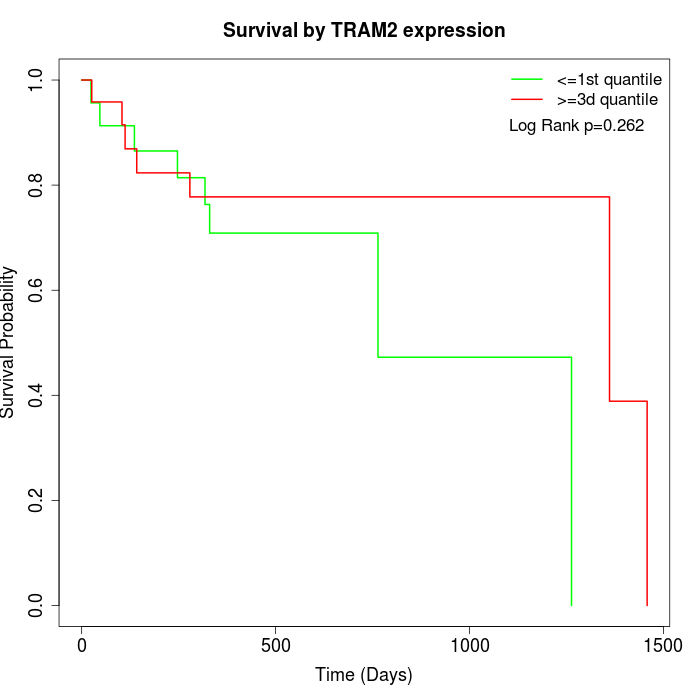
Survival by TRAM2 expression:
Note: Click image to view full size file.
Copy number change of TRAM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRAM2 | 9697 | 7 | 2 | 21 | |
| GSE20123 | TRAM2 | 9697 | 7 | 2 | 21 | |
| GSE43470 | TRAM2 | 9697 | 5 | 0 | 38 | |
| GSE46452 | TRAM2 | 9697 | 2 | 10 | 47 | |
| GSE47630 | TRAM2 | 9697 | 7 | 5 | 28 | |
| GSE54993 | TRAM2 | 9697 | 3 | 1 | 66 | |
| GSE54994 | TRAM2 | 9697 | 8 | 4 | 41 | |
| GSE60625 | TRAM2 | 9697 | 0 | 3 | 8 | |
| GSE74703 | TRAM2 | 9697 | 5 | 0 | 31 | |
| GSE74704 | TRAM2 | 9697 | 3 | 1 | 16 | |
| TCGA | TRAM2 | 9697 | 21 | 12 | 63 |
Total number of gains: 68; Total number of losses: 40; Total Number of normals: 380.
Somatic mutations of TRAM2:
Generating mutation plots.
Highly correlated genes for TRAM2:
Showing top 20/1944 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRAM2 | FCGR3A | 0.864451 | 3 | 0 | 3 |
| TRAM2 | WDR54 | 0.803297 | 7 | 0 | 7 |
| TRAM2 | LPCAT1 | 0.798677 | 11 | 0 | 11 |
| TRAM2 | CDH11 | 0.793488 | 13 | 0 | 13 |
| TRAM2 | PXDN | 0.792018 | 12 | 0 | 11 |
| TRAM2 | GNG10 | 0.773934 | 3 | 0 | 3 |
| TRAM2 | MFAP2 | 0.767773 | 12 | 0 | 11 |
| TRAM2 | VCAN | 0.766286 | 13 | 0 | 12 |
| TRAM2 | COL5A2 | 0.764121 | 12 | 0 | 12 |
| TRAM2 | LTBR | 0.758644 | 3 | 0 | 3 |
| TRAM2 | LOXL2 | 0.755669 | 12 | 0 | 11 |
| TRAM2 | FAM89A | 0.753814 | 3 | 0 | 3 |
| TRAM2 | SLC39A14 | 0.751244 | 13 | 0 | 13 |
| TRAM2 | CALU | 0.750326 | 12 | 0 | 12 |
| TRAM2 | CDK14 | 0.74663 | 10 | 0 | 9 |
| TRAM2 | TMEM263 | 0.745354 | 7 | 0 | 7 |
| TRAM2 | COL4A1 | 0.74428 | 13 | 0 | 12 |
| TRAM2 | SERPINH1 | 0.742327 | 11 | 0 | 10 |
| TRAM2 | PYCR2 | 0.741498 | 3 | 0 | 3 |
| TRAM2 | COL6A3 | 0.741183 | 13 | 0 | 13 |
For details and further investigation, click here