| Full name: LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated | Alias Symbol: G7b|YBL026W | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 57819 | HGNC ID: HGNC:13940 | Ensembl Gene: ENSG00000204392 | OMIM ID: 607282 |
| Drug and gene relationship at DGIdb | |||
Expression of LSM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LSM2 | 57819 | 209449_at | 0.7057 | 0.1184 | |
| GSE20347 | LSM2 | 57819 | 209449_at | 0.6920 | 0.0143 | |
| GSE23400 | LSM2 | 57819 | 209449_at | 0.7461 | 0.0000 | |
| GSE26886 | LSM2 | 57819 | 209449_at | 0.3175 | 0.4100 | |
| GSE29001 | LSM2 | 57819 | 209449_at | 0.6843 | 0.2005 | |
| GSE38129 | LSM2 | 57819 | 209449_at | 0.7332 | 0.0069 | |
| GSE45670 | LSM2 | 57819 | 209449_at | 0.3289 | 0.0410 | |
| GSE53622 | LSM2 | 57819 | 141237 | 0.6980 | 0.0000 | |
| GSE53624 | LSM2 | 57819 | 94190 | 0.5649 | 0.0000 | |
| GSE63941 | LSM2 | 57819 | 209449_at | 1.2130 | 0.0015 | |
| GSE77861 | LSM2 | 57819 | 209449_at | -0.0485 | 0.8688 | |
| GSE97050 | LSM2 | 57819 | A_23_P59153 | 0.4011 | 0.1976 | |
| SRP007169 | LSM2 | 57819 | RNAseq | 0.1456 | 0.6882 | |
| SRP008496 | LSM2 | 57819 | RNAseq | 0.1056 | 0.6824 | |
| SRP064894 | LSM2 | 57819 | RNAseq | 0.9470 | 0.0036 | |
| SRP133303 | LSM2 | 57819 | RNAseq | 0.3168 | 0.0133 | |
| SRP159526 | LSM2 | 57819 | RNAseq | 0.7179 | 0.0291 | |
| SRP193095 | LSM2 | 57819 | RNAseq | 0.1169 | 0.2842 | |
| SRP219564 | LSM2 | 57819 | RNAseq | 0.7938 | 0.0108 | |
| TCGA | LSM2 | 57819 | RNAseq | 0.2298 | 0.0001 |
Upregulated datasets: 1; Downregulated datasets: 0.
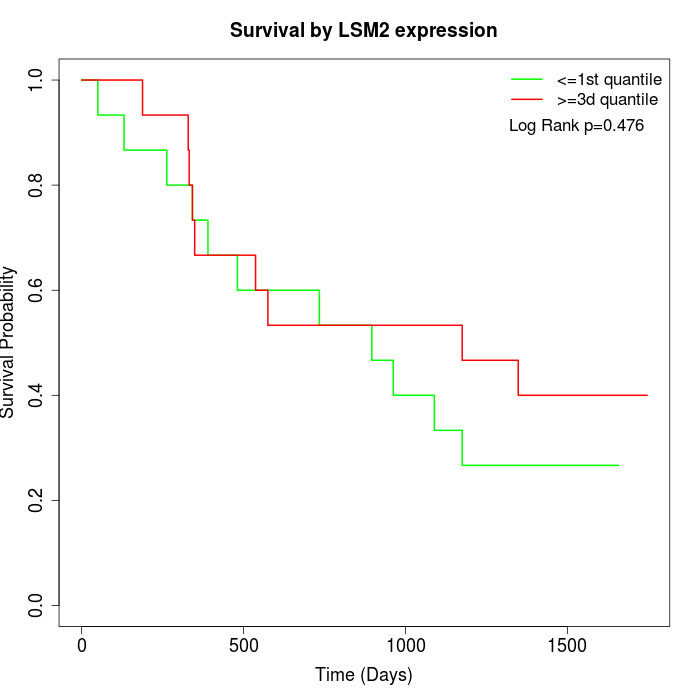
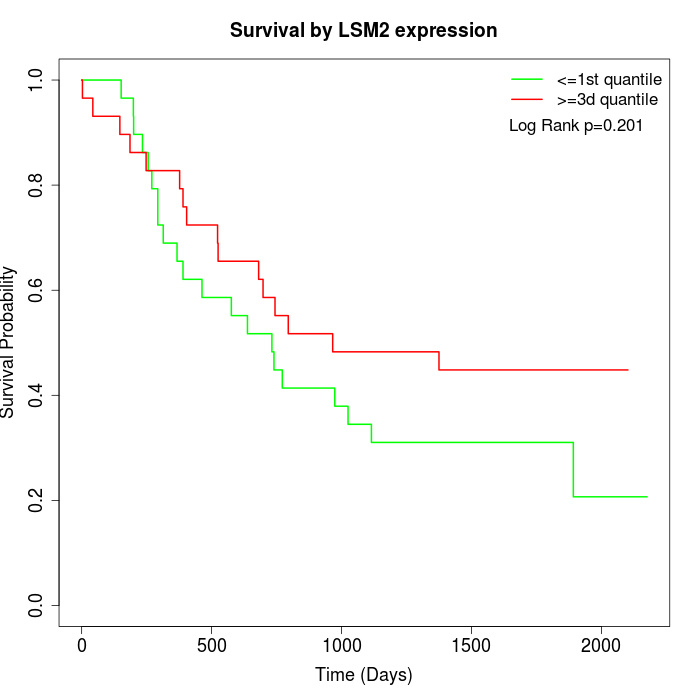
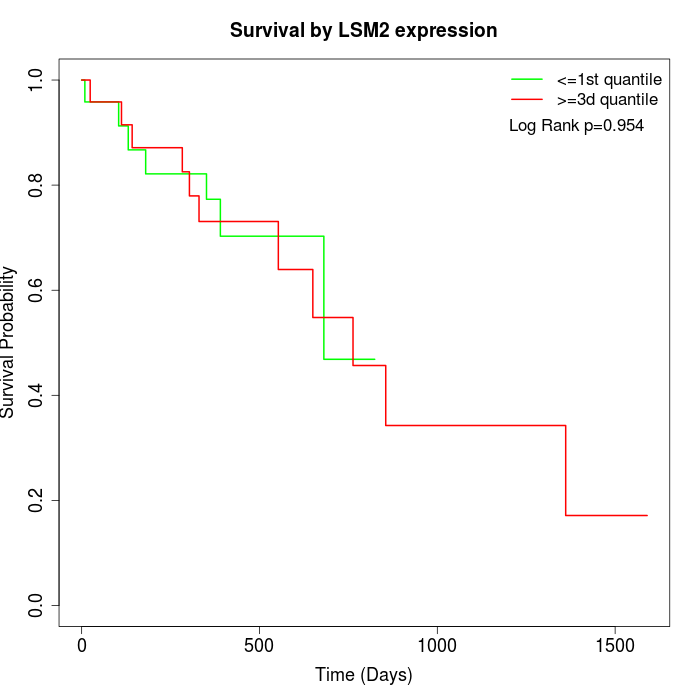
Survival by LSM2 expression:
Note: Click image to view full size file.
Copy number change of LSM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LSM2 | 57819 | 5 | 1 | 24 | |
| GSE20123 | LSM2 | 57819 | 5 | 1 | 24 | |
| GSE43470 | LSM2 | 57819 | 5 | 1 | 37 | |
| GSE46452 | LSM2 | 57819 | 1 | 10 | 48 | |
| GSE47630 | LSM2 | 57819 | 7 | 4 | 29 | |
| GSE54993 | LSM2 | 57819 | 2 | 1 | 67 | |
| GSE54994 | LSM2 | 57819 | 11 | 4 | 38 | |
| GSE60625 | LSM2 | 57819 | 0 | 1 | 10 | |
| GSE74703 | LSM2 | 57819 | 5 | 0 | 31 | |
| GSE74704 | LSM2 | 57819 | 2 | 0 | 18 | |
| TCGA | LSM2 | 57819 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 39; Total Number of normals: 390.
Somatic mutations of LSM2:
Generating mutation plots.
Highly correlated genes for LSM2:
Showing top 20/1743 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LSM2 | RAVER1 | 0.804361 | 3 | 0 | 3 |
| LSM2 | ELOF1 | 0.770407 | 3 | 0 | 3 |
| LSM2 | ZNF668 | 0.764205 | 4 | 0 | 4 |
| LSM2 | SCNM1 | 0.757746 | 3 | 0 | 3 |
| LSM2 | FBXO16 | 0.750888 | 3 | 0 | 3 |
| LSM2 | MRPL38 | 0.749821 | 4 | 0 | 3 |
| LSM2 | FTO | 0.749653 | 3 | 0 | 3 |
| LSM2 | SSR2 | 0.746479 | 10 | 0 | 10 |
| LSM2 | ABCF1 | 0.737137 | 12 | 0 | 11 |
| LSM2 | KDM1A | 0.736104 | 10 | 0 | 9 |
| LSM2 | RPRD1B | 0.731362 | 3 | 0 | 3 |
| LSM2 | RAB34 | 0.730154 | 3 | 0 | 3 |
| LSM2 | DNASE2 | 0.729263 | 4 | 0 | 4 |
| LSM2 | SNRPC | 0.729166 | 12 | 0 | 10 |
| LSM2 | CUTA | 0.724339 | 11 | 0 | 11 |
| LSM2 | HNRNPM | 0.718261 | 9 | 0 | 8 |
| LSM2 | SETD1A | 0.717112 | 5 | 0 | 4 |
| LSM2 | DGCR8 | 0.71094 | 3 | 0 | 3 |
| LSM2 | PFKM | 0.704715 | 5 | 0 | 5 |
| LSM2 | AXL | 0.704509 | 3 | 0 | 3 |
For details and further investigation, click here