| Full name: heterogeneous nuclear ribonucleoprotein M | Alias Symbol: HTGR1|HNRNPM4|HNRPM4|CEAR | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 4670 | HGNC ID: HGNC:5046 | Ensembl Gene: ENSG00000099783 | OMIM ID: 160994 |
| Drug and gene relationship at DGIdb | |||
Expression of HNRNPM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HNRNPM | 4670 | 200072_s_at | 0.3194 | 0.1193 | |
| GSE20347 | HNRNPM | 4670 | 200072_s_at | 0.5272 | 0.0004 | |
| GSE23400 | HNRNPM | 4670 | 200072_s_at | 0.5329 | 0.0000 | |
| GSE26886 | HNRNPM | 4670 | 1555844_s_at | -0.1450 | 0.4156 | |
| GSE29001 | HNRNPM | 4670 | 200072_s_at | 0.5695 | 0.1010 | |
| GSE38129 | HNRNPM | 4670 | 200072_s_at | 0.6209 | 0.0002 | |
| GSE45670 | HNRNPM | 4670 | 200072_s_at | 0.3576 | 0.0010 | |
| GSE53622 | HNRNPM | 4670 | 7611 | 0.2837 | 0.0000 | |
| GSE53624 | HNRNPM | 4670 | 7611 | 0.5155 | 0.0000 | |
| GSE63941 | HNRNPM | 4670 | 1555844_s_at | 0.1626 | 0.4095 | |
| GSE77861 | HNRNPM | 4670 | 1555844_s_at | 0.4930 | 0.0293 | |
| GSE97050 | HNRNPM | 4670 | A_33_P3242919 | 0.1220 | 0.5755 | |
| SRP007169 | HNRNPM | 4670 | RNAseq | 1.2877 | 0.0024 | |
| SRP008496 | HNRNPM | 4670 | RNAseq | 1.2791 | 0.0001 | |
| SRP064894 | HNRNPM | 4670 | RNAseq | 0.3422 | 0.0195 | |
| SRP133303 | HNRNPM | 4670 | RNAseq | 0.2516 | 0.0825 | |
| SRP159526 | HNRNPM | 4670 | RNAseq | 0.3433 | 0.0625 | |
| SRP193095 | HNRNPM | 4670 | RNAseq | 0.1698 | 0.0872 | |
| SRP219564 | HNRNPM | 4670 | RNAseq | 0.3158 | 0.2281 | |
| TCGA | HNRNPM | 4670 | RNAseq | 0.1034 | 0.0115 |
Upregulated datasets: 2; Downregulated datasets: 0.
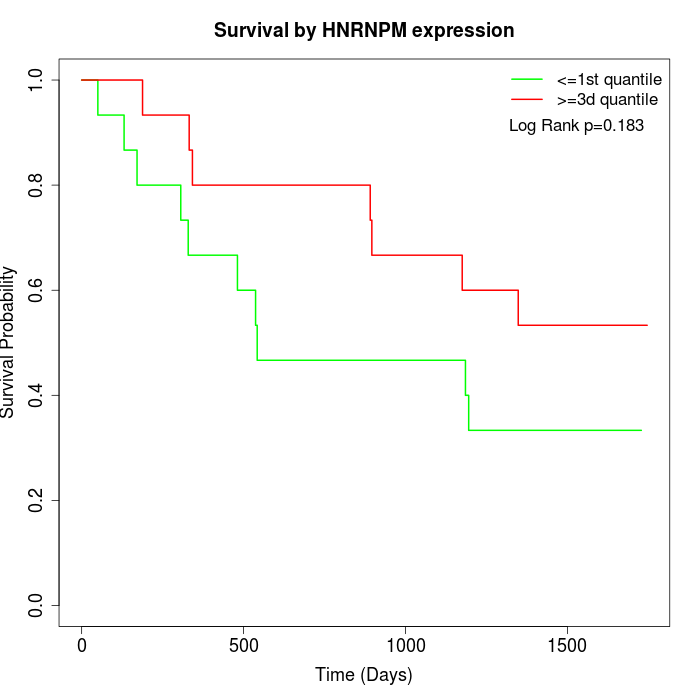
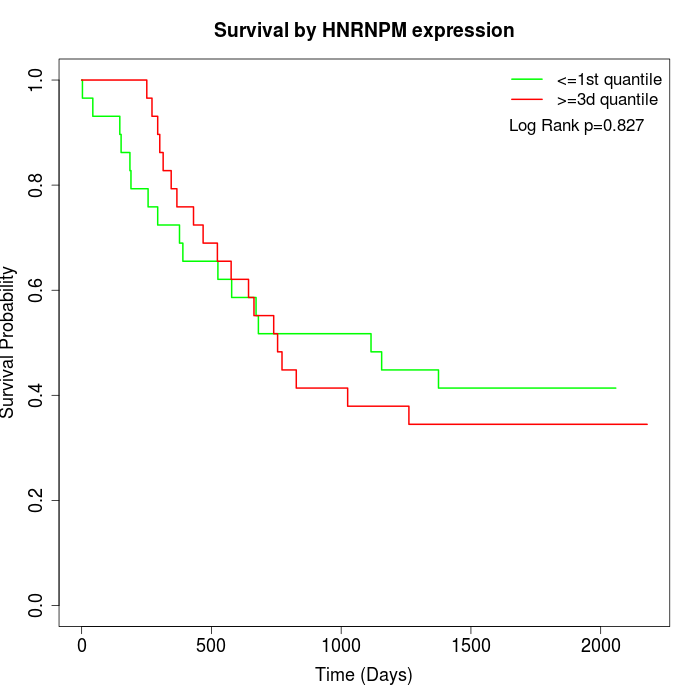
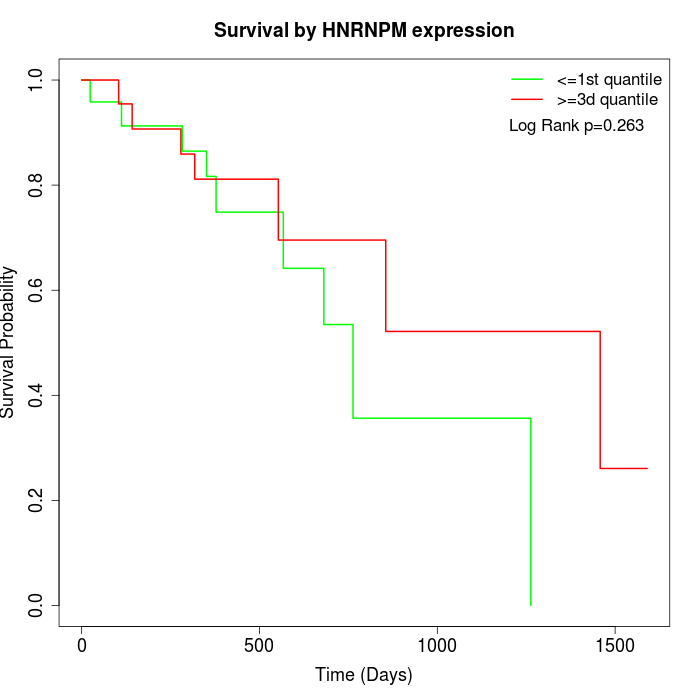
Survival by HNRNPM expression:
Note: Click image to view full size file.
Copy number change of HNRNPM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HNRNPM | 4670 | 4 | 3 | 23 | |
| GSE20123 | HNRNPM | 4670 | 3 | 2 | 25 | |
| GSE43470 | HNRNPM | 4670 | 2 | 6 | 35 | |
| GSE46452 | HNRNPM | 4670 | 47 | 1 | 11 | |
| GSE47630 | HNRNPM | 4670 | 5 | 7 | 28 | |
| GSE54993 | HNRNPM | 4670 | 16 | 3 | 51 | |
| GSE54994 | HNRNPM | 4670 | 6 | 14 | 33 | |
| GSE60625 | HNRNPM | 4670 | 9 | 0 | 2 | |
| GSE74703 | HNRNPM | 4670 | 2 | 4 | 30 | |
| GSE74704 | HNRNPM | 4670 | 0 | 2 | 18 | |
| TCGA | HNRNPM | 4670 | 10 | 19 | 67 |
Total number of gains: 104; Total number of losses: 61; Total Number of normals: 323.
Somatic mutations of HNRNPM:
Generating mutation plots.
Highly correlated genes for HNRNPM:
Showing top 20/1738 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HNRNPM | ZNF317 | 0.801784 | 4 | 0 | 4 |
| HNRNPM | RAB2B | 0.796381 | 3 | 0 | 3 |
| HNRNPM | C12orf73 | 0.794431 | 3 | 0 | 3 |
| HNRNPM | SLC39A3 | 0.755908 | 3 | 0 | 3 |
| HNRNPM | SMCR8 | 0.749206 | 4 | 0 | 3 |
| HNRNPM | GOT2 | 0.744953 | 3 | 0 | 3 |
| HNRNPM | NFATC3 | 0.740035 | 8 | 0 | 8 |
| HNRNPM | MRPS12 | 0.736764 | 9 | 0 | 9 |
| HNRNPM | CASK | 0.731798 | 5 | 0 | 5 |
| HNRNPM | ILF2 | 0.7312 | 11 | 0 | 11 |
| HNRNPM | MCOLN1 | 0.719718 | 4 | 0 | 4 |
| HNRNPM | ANKRD11 | 0.719325 | 3 | 0 | 3 |
| HNRNPM | LSM2 | 0.718261 | 9 | 0 | 8 |
| HNRNPM | FUS | 0.717831 | 12 | 0 | 11 |
| HNRNPM | SOCS4 | 0.715967 | 4 | 0 | 4 |
| HNRNPM | TSEN15 | 0.715631 | 5 | 0 | 4 |
| HNRNPM | TRPC4AP | 0.715372 | 3 | 0 | 3 |
| HNRNPM | EHMT2 | 0.712466 | 5 | 0 | 4 |
| HNRNPM | SNRPE | 0.711196 | 10 | 0 | 9 |
| HNRNPM | PELI2 | 0.709978 | 3 | 0 | 3 |
For details and further investigation, click here