| Full name: MAF bZIP transcription factor | Alias Symbol: c-MAF | ||
| Type: protein-coding gene | Cytoband: 16q23.2 | ||
| Entrez ID: 4094 | HGNC ID: HGNC:6776 | Ensembl Gene: ENSG00000178573 | OMIM ID: 177075 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAF involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05321 | Inflammatory bowel disease (IBD) |
Expression of MAF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAF | 4094 | 209348_s_at | -0.8476 | 0.2334 | |
| GSE20347 | MAF | 4094 | 209348_s_at | -0.9018 | 0.0100 | |
| GSE23400 | MAF | 4094 | 206363_at | 0.2257 | 0.0709 | |
| GSE26886 | MAF | 4094 | 209348_s_at | -1.9899 | 0.0001 | |
| GSE29001 | MAF | 4094 | 209348_s_at | -0.4885 | 0.3691 | |
| GSE38129 | MAF | 4094 | 209348_s_at | -0.5035 | 0.1035 | |
| GSE45670 | MAF | 4094 | 209348_s_at | 0.1320 | 0.6783 | |
| GSE53622 | MAF | 4094 | 31305 | -0.2070 | 0.2551 | |
| GSE53624 | MAF | 4094 | 31305 | -0.0972 | 0.4734 | |
| GSE63941 | MAF | 4094 | 209348_s_at | -2.6960 | 0.0376 | |
| GSE77861 | MAF | 4094 | 209348_s_at | -0.7906 | 0.3588 | |
| GSE97050 | MAF | 4094 | A_23_P397376 | 0.0939 | 0.8772 | |
| SRP007169 | MAF | 4094 | RNAseq | -1.4469 | 0.0080 | |
| SRP008496 | MAF | 4094 | RNAseq | -0.9870 | 0.0279 | |
| SRP064894 | MAF | 4094 | RNAseq | 0.1545 | 0.3248 | |
| SRP133303 | MAF | 4094 | RNAseq | -0.0096 | 0.9740 | |
| SRP159526 | MAF | 4094 | RNAseq | -0.3213 | 0.5418 | |
| SRP193095 | MAF | 4094 | RNAseq | -0.6312 | 0.0022 | |
| SRP219564 | MAF | 4094 | RNAseq | -0.2002 | 0.6851 | |
| TCGA | MAF | 4094 | RNAseq | 0.2631 | 0.0063 |
Upregulated datasets: 0; Downregulated datasets: 3.
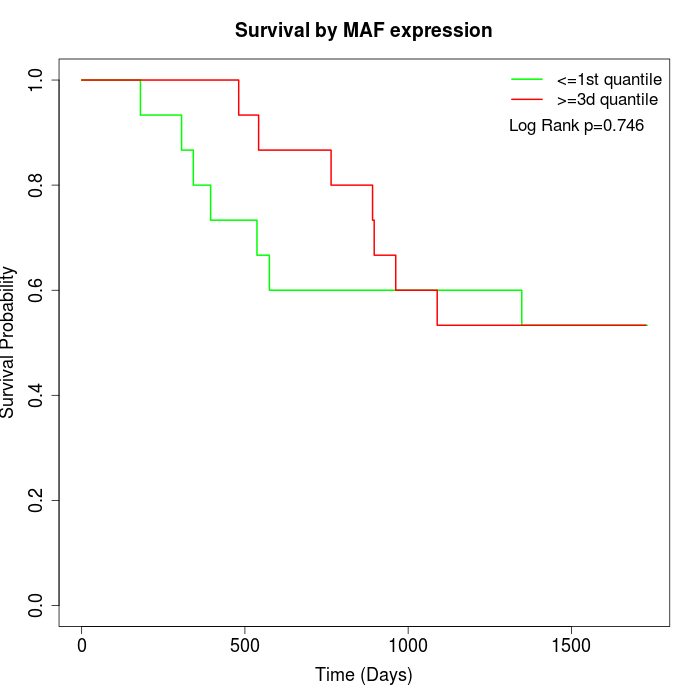
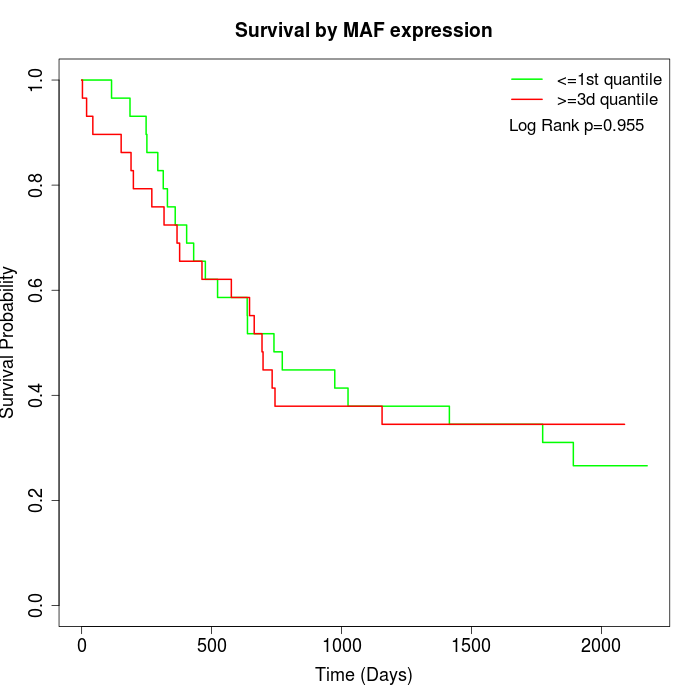
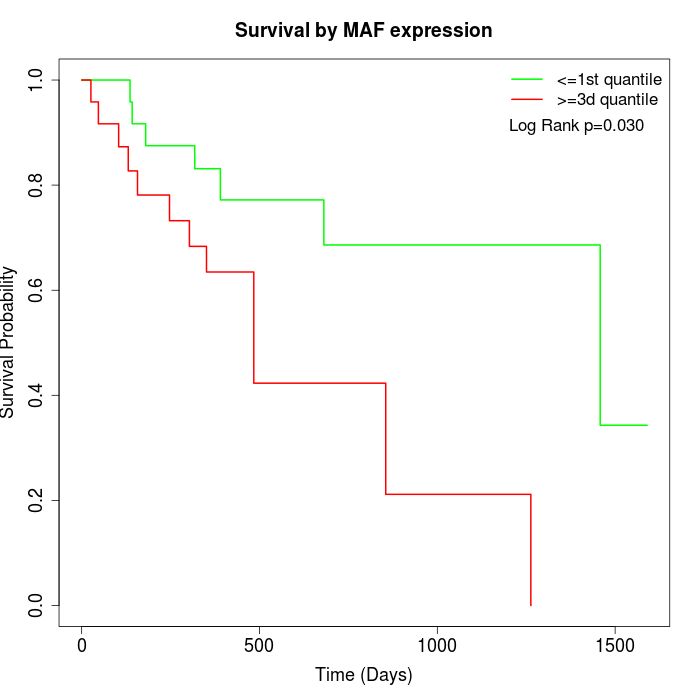
Survival by MAF expression:
Note: Click image to view full size file.
Copy number change of MAF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAF | 4094 | 4 | 3 | 23 | |
| GSE20123 | MAF | 4094 | 4 | 3 | 23 | |
| GSE43470 | MAF | 4094 | 2 | 8 | 33 | |
| GSE46452 | MAF | 4094 | 37 | 2 | 20 | |
| GSE47630 | MAF | 4094 | 11 | 8 | 21 | |
| GSE54993 | MAF | 4094 | 2 | 4 | 64 | |
| GSE54994 | MAF | 4094 | 7 | 10 | 36 | |
| GSE60625 | MAF | 4094 | 4 | 0 | 7 | |
| GSE74703 | MAF | 4094 | 2 | 6 | 28 | |
| GSE74704 | MAF | 4094 | 3 | 2 | 15 | |
| TCGA | MAF | 4094 | 24 | 15 | 57 |
Total number of gains: 100; Total number of losses: 61; Total Number of normals: 327.
Somatic mutations of MAF:
Generating mutation plots.
Highly correlated genes for MAF:
Showing top 20/335 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAF | ZCCHC9 | 0.703795 | 3 | 0 | 3 |
| MAF | IL20RB | 0.684676 | 5 | 0 | 5 |
| MAF | BNIPL | 0.671338 | 4 | 0 | 3 |
| MAF | MBOAT2 | 0.659531 | 5 | 0 | 3 |
| MAF | VSIG10L | 0.652951 | 4 | 0 | 3 |
| MAF | GJB6 | 0.649834 | 7 | 0 | 5 |
| MAF | LYPD3 | 0.643682 | 8 | 0 | 7 |
| MAF | CETN3 | 0.640317 | 3 | 0 | 3 |
| MAF | PRKAR1A | 0.638227 | 5 | 0 | 5 |
| MAF | S100A12 | 0.633358 | 6 | 0 | 5 |
| MAF | FAM174A | 0.629097 | 4 | 0 | 3 |
| MAF | LEPROT | 0.627235 | 3 | 0 | 3 |
| MAF | LYSMD3 | 0.620589 | 5 | 0 | 4 |
| MAF | XG | 0.619382 | 5 | 0 | 4 |
| MAF | GSKIP | 0.617136 | 4 | 0 | 3 |
| MAF | CLCA2 | 0.617089 | 9 | 0 | 8 |
| MAF | PTGR1 | 0.614613 | 4 | 0 | 3 |
| MAF | PLIN3 | 0.609373 | 8 | 0 | 6 |
| MAF | IDH2 | 0.609219 | 5 | 0 | 5 |
| MAF | SMIM15 | 0.606495 | 3 | 0 | 3 |
For details and further investigation, click here