| Full name: mitogen-activated protein kinase kinase kinase 2 | Alias Symbol: MEKK2B | ||
| Type: protein-coding gene | Cytoband: 2q14.3 | ||
| Entrez ID: 10746 | HGNC ID: HGNC:6854 | Ensembl Gene: ENSG00000169967 | OMIM ID: 609487 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAP3K2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04540 | Gap junction | |
| hsa04912 | GnRH signaling pathway |
Expression of MAP3K2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP3K2 | 10746 | 226979_at | 0.6675 | 0.0250 | |
| GSE20347 | MAP3K2 | 10746 | 221695_s_at | 0.1712 | 0.1224 | |
| GSE23400 | MAP3K2 | 10746 | 221695_s_at | 0.2678 | 0.0000 | |
| GSE26886 | MAP3K2 | 10746 | 226979_at | 1.6026 | 0.0000 | |
| GSE29001 | MAP3K2 | 10746 | 221695_s_at | 0.1793 | 0.0922 | |
| GSE38129 | MAP3K2 | 10746 | 221695_s_at | 0.2236 | 0.0482 | |
| GSE45670 | MAP3K2 | 10746 | 226979_at | 0.4049 | 0.0040 | |
| GSE53622 | MAP3K2 | 10746 | 69616 | 0.5903 | 0.0000 | |
| GSE53624 | MAP3K2 | 10746 | 69616 | 0.3348 | 0.0000 | |
| GSE63941 | MAP3K2 | 10746 | 226979_at | 0.6004 | 0.1163 | |
| GSE77861 | MAP3K2 | 10746 | 226979_at | 0.9562 | 0.0023 | |
| GSE97050 | MAP3K2 | 10746 | A_33_P3415445 | 0.4881 | 0.1403 | |
| SRP007169 | MAP3K2 | 10746 | RNAseq | 0.8893 | 0.0117 | |
| SRP008496 | MAP3K2 | 10746 | RNAseq | 0.9974 | 0.0000 | |
| SRP064894 | MAP3K2 | 10746 | RNAseq | 0.4364 | 0.0570 | |
| SRP133303 | MAP3K2 | 10746 | RNAseq | 0.7155 | 0.0000 | |
| SRP159526 | MAP3K2 | 10746 | RNAseq | 0.1930 | 0.2678 | |
| SRP193095 | MAP3K2 | 10746 | RNAseq | 0.2197 | 0.0672 | |
| SRP219564 | MAP3K2 | 10746 | RNAseq | 0.1122 | 0.6409 | |
| TCGA | MAP3K2 | 10746 | RNAseq | 0.0150 | 0.8098 |
Upregulated datasets: 1; Downregulated datasets: 0.
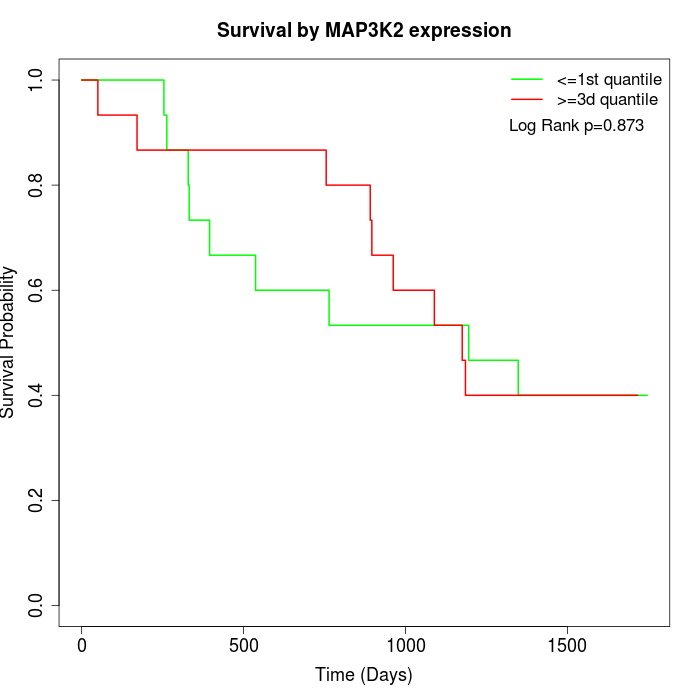
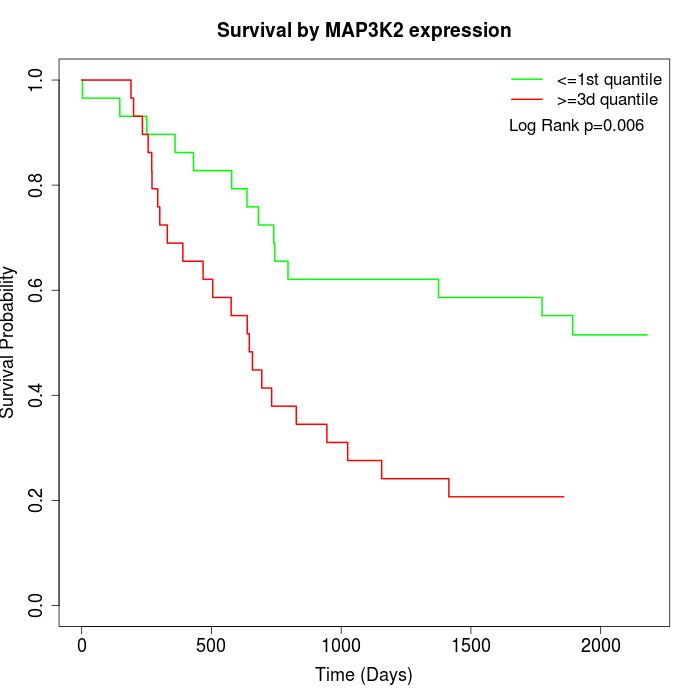
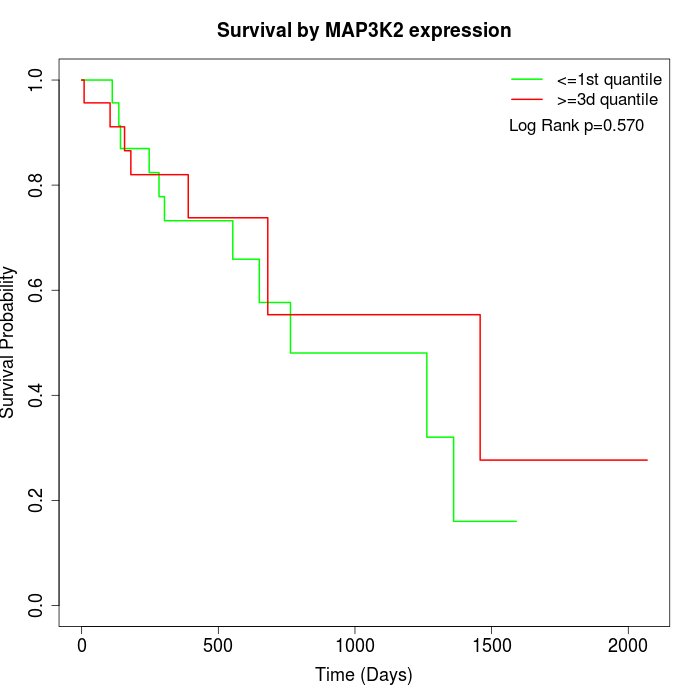
Survival by MAP3K2 expression:
Note: Click image to view full size file.
Copy number change of MAP3K2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP3K2 | 10746 | 5 | 3 | 22 | |
| GSE20123 | MAP3K2 | 10746 | 6 | 3 | 21 | |
| GSE43470 | MAP3K2 | 10746 | 2 | 1 | 40 | |
| GSE46452 | MAP3K2 | 10746 | 1 | 3 | 55 | |
| GSE47630 | MAP3K2 | 10746 | 6 | 0 | 34 | |
| GSE54993 | MAP3K2 | 10746 | 0 | 6 | 64 | |
| GSE54994 | MAP3K2 | 10746 | 11 | 0 | 42 | |
| GSE60625 | MAP3K2 | 10746 | 0 | 3 | 8 | |
| GSE74703 | MAP3K2 | 10746 | 2 | 1 | 33 | |
| GSE74704 | MAP3K2 | 10746 | 3 | 1 | 16 | |
| TCGA | MAP3K2 | 10746 | 22 | 8 | 66 |
Total number of gains: 58; Total number of losses: 29; Total Number of normals: 401.
Somatic mutations of MAP3K2:
Generating mutation plots.
Highly correlated genes for MAP3K2:
Showing top 20/1675 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP3K2 | GNS | 0.821995 | 3 | 0 | 3 |
| MAP3K2 | ZCCHC7 | 0.817209 | 3 | 0 | 3 |
| MAP3K2 | RNF4 | 0.785144 | 3 | 0 | 3 |
| MAP3K2 | WDR4 | 0.784434 | 3 | 0 | 3 |
| MAP3K2 | EIF3M | 0.77376 | 3 | 0 | 3 |
| MAP3K2 | YWHAG | 0.751733 | 5 | 0 | 5 |
| MAP3K2 | SPIRE1 | 0.750045 | 3 | 0 | 3 |
| MAP3K2 | ZNF451 | 0.744432 | 4 | 0 | 4 |
| MAP3K2 | MORC2 | 0.744101 | 8 | 0 | 7 |
| MAP3K2 | TMEM184B | 0.739121 | 7 | 0 | 7 |
| MAP3K2 | GEMIN5 | 0.738946 | 3 | 0 | 3 |
| MAP3K2 | ZNF627 | 0.738018 | 3 | 0 | 3 |
| MAP3K2 | SLC25A28 | 0.735486 | 3 | 0 | 3 |
| MAP3K2 | JMJD4 | 0.734964 | 3 | 0 | 3 |
| MAP3K2 | PSMG3 | 0.734439 | 5 | 0 | 5 |
| MAP3K2 | TMEM138 | 0.729925 | 7 | 0 | 7 |
| MAP3K2 | MRPS26 | 0.729619 | 4 | 0 | 3 |
| MAP3K2 | HNRNPUL1 | 0.728081 | 3 | 0 | 3 |
| MAP3K2 | TTL | 0.727005 | 6 | 0 | 5 |
| MAP3K2 | CEP170 | 0.723893 | 3 | 0 | 3 |
For details and further investigation, click here