| Full name: MAPK interacting serine/threonine kinase 2 | Alias Symbol: MNK2 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 2872 | HGNC ID: HGNC:7111 | Ensembl Gene: ENSG00000099875 | OMIM ID: 605069 |
| Drug and gene relationship at DGIdb | |||
MKNK2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04066 | HIF-1 signaling pathway | |
| hsa04910 | Insulin signaling pathway |
Expression of MKNK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MKNK2 | 2872 | 218205_s_at | -0.8409 | 0.0282 | |
| GSE20347 | MKNK2 | 2872 | 218205_s_at | -0.6422 | 0.0045 | |
| GSE23400 | MKNK2 | 2872 | 218205_s_at | -0.7026 | 0.0000 | |
| GSE26886 | MKNK2 | 2872 | 218205_s_at | -0.5907 | 0.0101 | |
| GSE29001 | MKNK2 | 2872 | 218205_s_at | -0.8780 | 0.0028 | |
| GSE38129 | MKNK2 | 2872 | 218205_s_at | -0.6056 | 0.0014 | |
| GSE45670 | MKNK2 | 2872 | 218205_s_at | -0.3654 | 0.0751 | |
| GSE53622 | MKNK2 | 2872 | 41982 | -1.5981 | 0.0000 | |
| GSE53624 | MKNK2 | 2872 | 41982 | -1.2987 | 0.0000 | |
| GSE63941 | MKNK2 | 2872 | 218205_s_at | 0.0296 | 0.9624 | |
| GSE77861 | MKNK2 | 2872 | 218205_s_at | -0.6714 | 0.1055 | |
| GSE97050 | MKNK2 | 2872 | A_23_P142310 | -1.0693 | 0.1078 | |
| SRP007169 | MKNK2 | 2872 | RNAseq | -2.7843 | 0.0000 | |
| SRP008496 | MKNK2 | 2872 | RNAseq | -2.8353 | 0.0000 | |
| SRP064894 | MKNK2 | 2872 | RNAseq | -1.5805 | 0.0000 | |
| SRP133303 | MKNK2 | 2872 | RNAseq | -1.1951 | 0.0000 | |
| SRP159526 | MKNK2 | 2872 | RNAseq | -1.2597 | 0.0032 | |
| SRP193095 | MKNK2 | 2872 | RNAseq | -1.2401 | 0.0000 | |
| SRP219564 | MKNK2 | 2872 | RNAseq | -1.7125 | 0.0001 | |
| TCGA | MKNK2 | 2872 | RNAseq | -0.1732 | 0.0034 |
Upregulated datasets: 0; Downregulated datasets: 9.
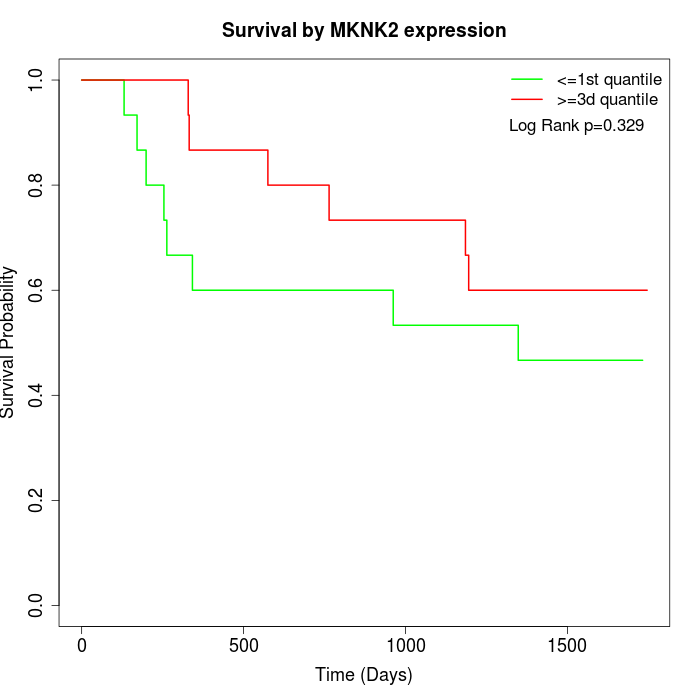
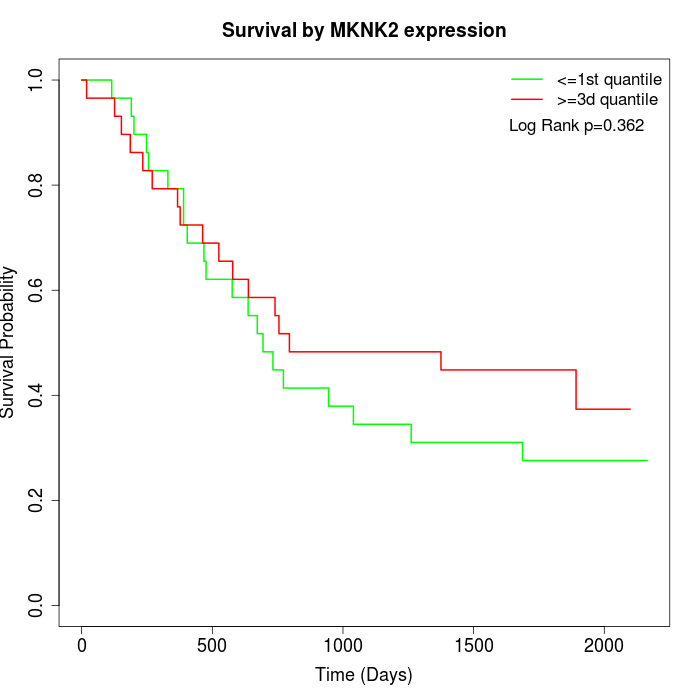
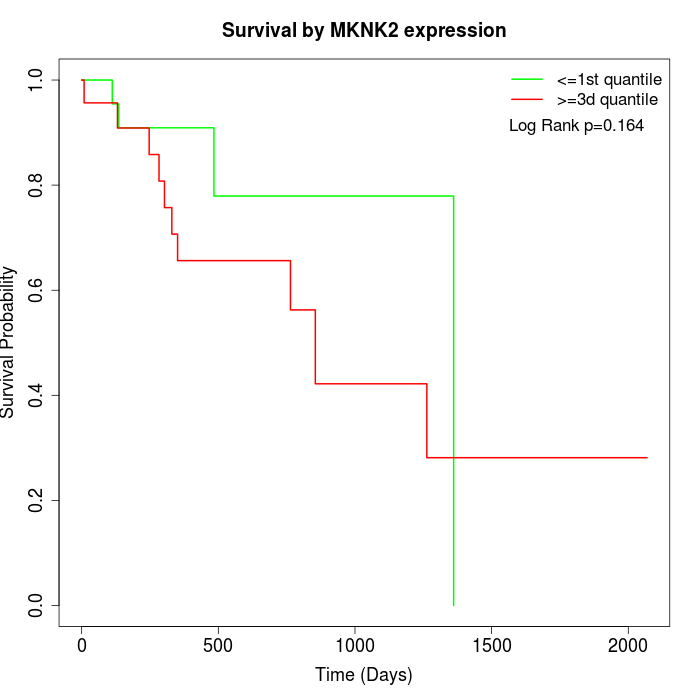
Survival by MKNK2 expression:
Note: Click image to view full size file.
Copy number change of MKNK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MKNK2 | 2872 | 4 | 6 | 20 | |
| GSE20123 | MKNK2 | 2872 | 4 | 5 | 21 | |
| GSE43470 | MKNK2 | 2872 | 1 | 10 | 32 | |
| GSE46452 | MKNK2 | 2872 | 47 | 1 | 11 | |
| GSE47630 | MKNK2 | 2872 | 5 | 7 | 28 | |
| GSE54993 | MKNK2 | 2872 | 16 | 3 | 51 | |
| GSE54994 | MKNK2 | 2872 | 8 | 15 | 30 | |
| GSE60625 | MKNK2 | 2872 | 9 | 0 | 2 | |
| GSE74703 | MKNK2 | 2872 | 1 | 7 | 28 | |
| GSE74704 | MKNK2 | 2872 | 3 | 3 | 14 | |
| TCGA | MKNK2 | 2872 | 6 | 24 | 66 |
Total number of gains: 104; Total number of losses: 81; Total Number of normals: 303.
Somatic mutations of MKNK2:
Generating mutation plots.
Highly correlated genes for MKNK2:
Showing top 20/1508 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MKNK2 | BOK | 0.761406 | 5 | 0 | 5 |
| MKNK2 | ZDHHC21 | 0.752113 | 4 | 0 | 4 |
| MKNK2 | TMEM38A | 0.748869 | 3 | 0 | 3 |
| MKNK2 | DDI2 | 0.742726 | 3 | 0 | 3 |
| MKNK2 | SMIM5 | 0.739088 | 6 | 0 | 6 |
| MKNK2 | CASC3 | 0.737292 | 3 | 0 | 3 |
| MKNK2 | SLC10A5 | 0.728509 | 3 | 0 | 3 |
| MKNK2 | SMDT1 | 0.727636 | 3 | 0 | 3 |
| MKNK2 | RSPH3 | 0.725343 | 4 | 0 | 4 |
| MKNK2 | CA13 | 0.723342 | 3 | 0 | 3 |
| MKNK2 | TRIP10 | 0.723334 | 12 | 0 | 12 |
| MKNK2 | MORN3 | 0.721564 | 3 | 0 | 3 |
| MKNK2 | PHYHD1 | 0.718934 | 6 | 0 | 6 |
| MKNK2 | ZDHHC5 | 0.717627 | 3 | 0 | 3 |
| MKNK2 | ATP6V1G1 | 0.71646 | 3 | 0 | 3 |
| MKNK2 | EPGN | 0.708071 | 3 | 0 | 3 |
| MKNK2 | ATP8B1 | 0.707252 | 3 | 0 | 3 |
| MKNK2 | GID4 | 0.703658 | 5 | 0 | 5 |
| MKNK2 | SLC25A27 | 0.699063 | 3 | 0 | 3 |
| MKNK2 | C4orf3 | 0.698604 | 7 | 0 | 7 |
For details and further investigation, click here