| Full name: MAX dimerization protein MLX | Alias Symbol: MAD7|MXD7|bHLHd13 | ||
| Type: protein-coding gene | Cytoband: 17q21.1 | ||
| Entrez ID: 6945 | HGNC ID: HGNC:11645 | Ensembl Gene: ENSG00000108788 | OMIM ID: 602976 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MLX involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04931 | Insulin resistance | |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) |
Expression of MLX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MLX | 6945 | 213708_s_at | 0.0068 | 0.9883 | |
| GSE20347 | MLX | 6945 | 213708_s_at | -0.2473 | 0.0460 | |
| GSE23400 | MLX | 6945 | 213708_s_at | 0.0531 | 0.2476 | |
| GSE26886 | MLX | 6945 | 213708_s_at | -0.1498 | 0.2562 | |
| GSE29001 | MLX | 6945 | 213708_s_at | -0.4127 | 0.0035 | |
| GSE38129 | MLX | 6945 | 213708_s_at | -0.1558 | 0.2175 | |
| GSE45670 | MLX | 6945 | 213708_s_at | 0.0839 | 0.5111 | |
| GSE53622 | MLX | 6945 | 63243 | -0.1741 | 0.0067 | |
| GSE53624 | MLX | 6945 | 20516 | 0.0518 | 0.4046 | |
| GSE63941 | MLX | 6945 | 213708_s_at | 0.4621 | 0.0887 | |
| GSE77861 | MLX | 6945 | 213708_s_at | 0.0646 | 0.7183 | |
| GSE97050 | MLX | A_19_P00810045 | -0.0367 | 0.8776 | ||
| SRP007169 | MLX | 6945 | RNAseq | -0.2409 | 0.5074 | |
| SRP008496 | MLX | 6945 | RNAseq | -0.3108 | 0.1399 | |
| SRP064894 | MLX | 6945 | RNAseq | 0.0230 | 0.8554 | |
| SRP133303 | MLX | 6945 | RNAseq | 0.1060 | 0.4712 | |
| SRP159526 | MLX | 6945 | RNAseq | -0.0631 | 0.8030 | |
| SRP193095 | MLX | 6945 | RNAseq | -0.3814 | 0.0004 | |
| SRP219564 | MLX | 6945 | RNAseq | -0.1962 | 0.5519 | |
| TCGA | MLX | 6945 | RNAseq | -0.0600 | 0.2041 |
Upregulated datasets: 0; Downregulated datasets: 0.
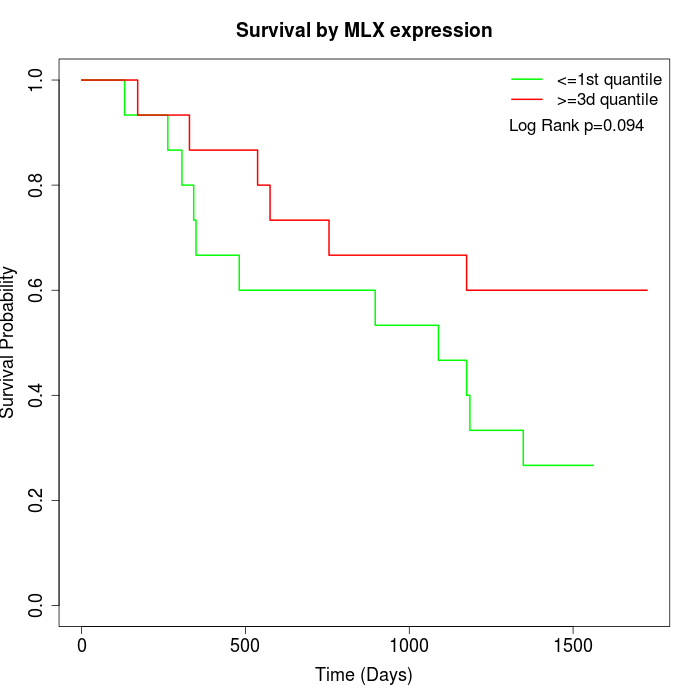
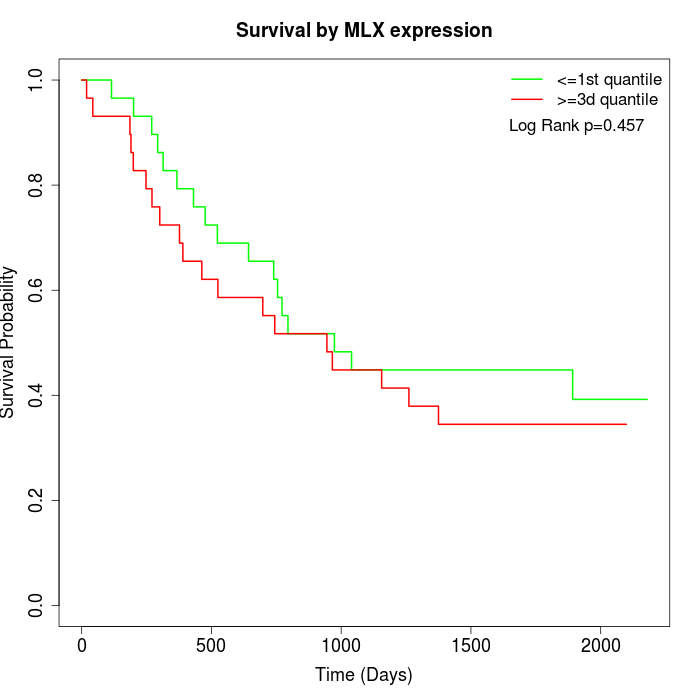
Survival by MLX expression:
Note: Click image to view full size file.
Copy number change of MLX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MLX | 6945 | 6 | 2 | 22 | |
| GSE20123 | MLX | 6945 | 6 | 2 | 22 | |
| GSE43470 | MLX | 6945 | 1 | 2 | 40 | |
| GSE46452 | MLX | 6945 | 34 | 0 | 25 | |
| GSE47630 | MLX | 6945 | 8 | 1 | 31 | |
| GSE54993 | MLX | 6945 | 3 | 4 | 63 | |
| GSE54994 | MLX | 6945 | 8 | 5 | 40 | |
| GSE60625 | MLX | 6945 | 4 | 0 | 7 | |
| GSE74703 | MLX | 6945 | 1 | 1 | 34 | |
| GSE74704 | MLX | 6945 | 4 | 1 | 15 | |
| TCGA | MLX | 6945 | 23 | 6 | 67 |
Total number of gains: 98; Total number of losses: 24; Total Number of normals: 366.
Somatic mutations of MLX:
Generating mutation plots.
Highly correlated genes for MLX:
Showing top 20/218 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MLX | LRRC28 | 0.720048 | 3 | 0 | 3 |
| MLX | MCFD2 | 0.712682 | 3 | 0 | 3 |
| MLX | TNFAIP2 | 0.692658 | 3 | 0 | 3 |
| MLX | PIGS | 0.667787 | 4 | 0 | 4 |
| MLX | DNPEP | 0.665881 | 3 | 0 | 3 |
| MLX | ZNF594 | 0.66455 | 3 | 0 | 3 |
| MLX | GRIN3B | 0.663025 | 3 | 0 | 3 |
| MLX | ATP6V0E1 | 0.656715 | 5 | 0 | 4 |
| MLX | MCTP2 | 0.644348 | 3 | 0 | 3 |
| MLX | MAPK1 | 0.641845 | 3 | 0 | 3 |
| MLX | THAP2 | 0.641543 | 3 | 0 | 3 |
| MLX | VPS33A | 0.640976 | 3 | 0 | 3 |
| MLX | FAM162B | 0.627169 | 3 | 0 | 3 |
| MLX | OR2F1 | 0.624513 | 4 | 0 | 3 |
| MLX | PPP1R16B | 0.623922 | 4 | 0 | 4 |
| MLX | SERPINB8 | 0.618464 | 5 | 0 | 3 |
| MLX | MC2R | 0.617766 | 3 | 0 | 3 |
| MLX | NRBF2 | 0.616121 | 4 | 0 | 3 |
| MLX | HOXB4 | 0.615855 | 3 | 0 | 3 |
| MLX | ACAD9 | 0.615607 | 3 | 0 | 3 |
For details and further investigation, click here