| Full name: ATPase H+ transporting V0 subunit e1 | Alias Symbol: M9.2 | ||
| Type: protein-coding gene | Cytoband: 5q35.1 | ||
| Entrez ID: 8992 | HGNC ID: HGNC:863 | Ensembl Gene: ENSG00000113732 | OMIM ID: 603931 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V0E1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V0E1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0E1 | 8992 | 200096_s_at | -0.0063 | 0.9892 | |
| GSE20347 | ATP6V0E1 | 8992 | 214150_x_at | -0.1816 | 0.0500 | |
| GSE23400 | ATP6V0E1 | 8992 | 201172_x_at | -0.1685 | 0.0087 | |
| GSE26886 | ATP6V0E1 | 8992 | 214150_x_at | -0.0766 | 0.5482 | |
| GSE29001 | ATP6V0E1 | 8992 | 214150_x_at | -0.2067 | 0.0525 | |
| GSE38129 | ATP6V0E1 | 8992 | 214150_x_at | -0.2090 | 0.0087 | |
| GSE45670 | ATP6V0E1 | 8992 | 200096_s_at | -0.1726 | 0.2244 | |
| GSE53622 | ATP6V0E1 | 8992 | 40760 | 0.0497 | 0.4580 | |
| GSE53624 | ATP6V0E1 | 8992 | 40760 | -0.0452 | 0.3701 | |
| GSE63941 | ATP6V0E1 | 8992 | 200096_s_at | -0.5302 | 0.1665 | |
| GSE77861 | ATP6V0E1 | 8992 | 201172_x_at | -0.2318 | 0.3576 | |
| GSE97050 | ATP6V0E1 | 8992 | A_23_P213840 | -0.0200 | 0.9633 | |
| SRP007169 | ATP6V0E1 | 8992 | RNAseq | -1.7516 | 0.0000 | |
| SRP008496 | ATP6V0E1 | 8992 | RNAseq | -1.6790 | 0.0000 | |
| SRP064894 | ATP6V0E1 | 8992 | RNAseq | -0.1862 | 0.4705 | |
| SRP133303 | ATP6V0E1 | 8992 | RNAseq | 0.0612 | 0.7574 | |
| SRP159526 | ATP6V0E1 | 8992 | RNAseq | -0.3535 | 0.3974 | |
| SRP193095 | ATP6V0E1 | 8992 | RNAseq | -0.4869 | 0.0022 | |
| SRP219564 | ATP6V0E1 | 8992 | RNAseq | -0.2130 | 0.4151 | |
| TCGA | ATP6V0E1 | 8992 | RNAseq | 0.0234 | 0.6224 |
Upregulated datasets: 0; Downregulated datasets: 2.
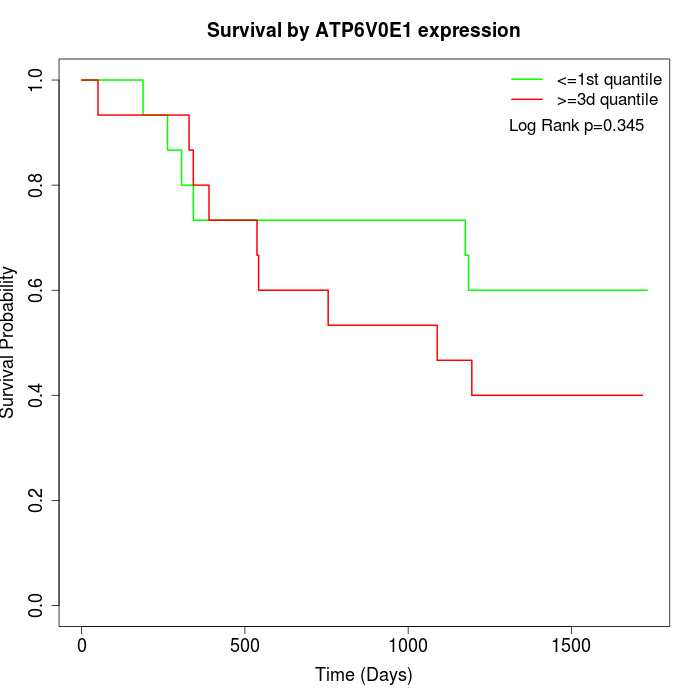
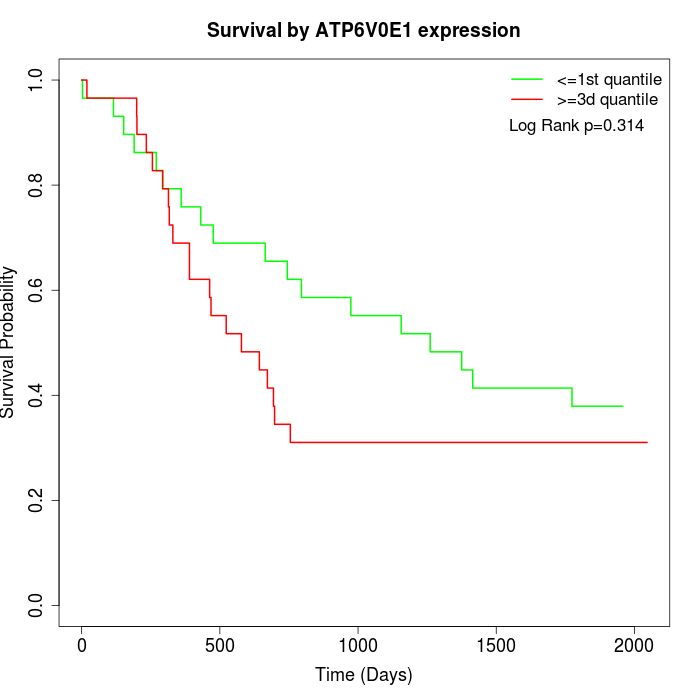
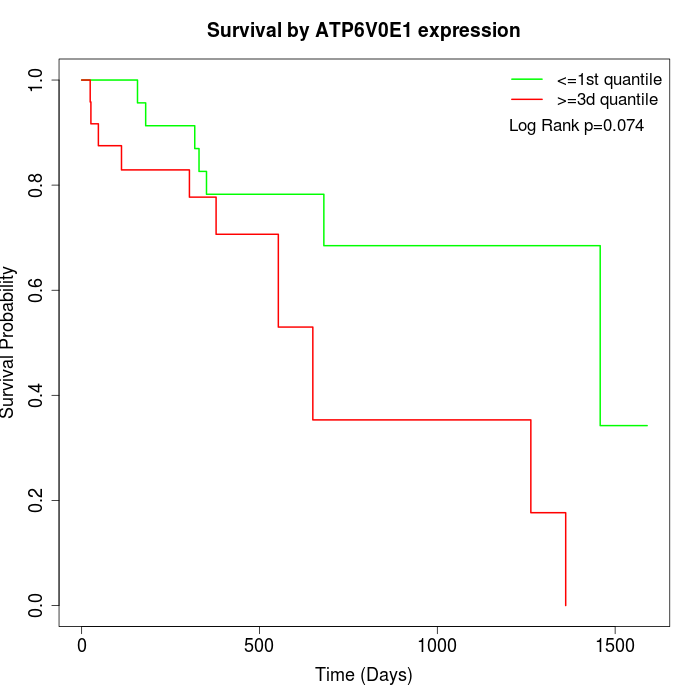
Survival by ATP6V0E1 expression:
Note: Click image to view full size file.
Copy number change of ATP6V0E1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0E1 | 8992 | 2 | 12 | 16 | |
| GSE20123 | ATP6V0E1 | 8992 | 2 | 12 | 16 | |
| GSE43470 | ATP6V0E1 | 8992 | 2 | 10 | 31 | |
| GSE46452 | ATP6V0E1 | 8992 | 0 | 27 | 32 | |
| GSE47630 | ATP6V0E1 | 8992 | 0 | 20 | 20 | |
| GSE54993 | ATP6V0E1 | 8992 | 9 | 2 | 59 | |
| GSE54994 | ATP6V0E1 | 8992 | 2 | 15 | 36 | |
| GSE60625 | ATP6V0E1 | 8992 | 1 | 0 | 10 | |
| GSE74703 | ATP6V0E1 | 8992 | 2 | 7 | 27 | |
| GSE74704 | ATP6V0E1 | 8992 | 1 | 6 | 13 | |
| TCGA | ATP6V0E1 | 8992 | 8 | 34 | 54 |
Total number of gains: 29; Total number of losses: 145; Total Number of normals: 314.
Somatic mutations of ATP6V0E1:
Generating mutation plots.
Highly correlated genes for ATP6V0E1:
Showing top 20/362 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0E1 | MVP | 0.80812 | 3 | 0 | 3 |
| ATP6V0E1 | CELA3A | 0.754409 | 3 | 0 | 3 |
| ATP6V0E1 | HINT3 | 0.740299 | 3 | 0 | 3 |
| ATP6V0E1 | PRELID1 | 0.712858 | 4 | 0 | 4 |
| ATP6V0E1 | PTGR1 | 0.707877 | 3 | 0 | 3 |
| ATP6V0E1 | NRBF2 | 0.707171 | 4 | 0 | 3 |
| ATP6V0E1 | GNAI3 | 0.702739 | 3 | 0 | 3 |
| ATP6V0E1 | LRRD1 | 0.694245 | 3 | 0 | 3 |
| ATP6V0E1 | CPEB4 | 0.690968 | 4 | 0 | 4 |
| ATP6V0E1 | SMG8 | 0.689785 | 3 | 0 | 3 |
| ATP6V0E1 | HEXIM1 | 0.684702 | 3 | 0 | 3 |
| ATP6V0E1 | APBA1 | 0.673813 | 3 | 0 | 3 |
| ATP6V0E1 | VPS25 | 0.672971 | 4 | 0 | 3 |
| ATP6V0E1 | MRPL30 | 0.672723 | 3 | 0 | 3 |
| ATP6V0E1 | SELP | 0.670277 | 3 | 0 | 3 |
| ATP6V0E1 | SCPEP1 | 0.669442 | 3 | 0 | 3 |
| ATP6V0E1 | OSTC | 0.666494 | 3 | 0 | 3 |
| ATP6V0E1 | ZDHHC21 | 0.665572 | 3 | 0 | 3 |
| ATP6V0E1 | COPZ1 | 0.665152 | 4 | 0 | 3 |
| ATP6V0E1 | CMTR2 | 0.661478 | 4 | 0 | 3 |
For details and further investigation, click here