| Full name: N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | Alias Symbol: APAA|UCE | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 51172 | HGNC ID: HGNC:17378 | Ensembl Gene: ENSG00000103174 | OMIM ID: 607985 |
| Drug and gene relationship at DGIdb | |||
Expression of NAGPA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAGPA | 51172 | 205090_s_at | 0.4277 | 0.2223 | |
| GSE20347 | NAGPA | 51172 | 205090_s_at | 0.2606 | 0.0413 | |
| GSE23400 | NAGPA | 51172 | 205090_s_at | 0.1907 | 0.0001 | |
| GSE26886 | NAGPA | 51172 | 205090_s_at | 0.3588 | 0.0707 | |
| GSE29001 | NAGPA | 51172 | 205090_s_at | 0.2942 | 0.0449 | |
| GSE38129 | NAGPA | 51172 | 205090_s_at | 0.2455 | 0.0266 | |
| GSE45670 | NAGPA | 51172 | 205090_s_at | 0.2853 | 0.0055 | |
| GSE53622 | NAGPA | 51172 | 14021 | 0.5089 | 0.0000 | |
| GSE53624 | NAGPA | 51172 | 14021 | 0.4349 | 0.0000 | |
| GSE63941 | NAGPA | 51172 | 205090_s_at | -0.5889 | 0.0842 | |
| GSE77861 | NAGPA | 51172 | 205090_s_at | 0.2012 | 0.3592 | |
| GSE97050 | NAGPA | 51172 | A_23_P38041 | 0.2570 | 0.2650 | |
| SRP007169 | NAGPA | 51172 | RNAseq | 0.9112 | 0.0631 | |
| SRP064894 | NAGPA | 51172 | RNAseq | 0.7455 | 0.0009 | |
| SRP133303 | NAGPA | 51172 | RNAseq | 0.3048 | 0.1400 | |
| SRP159526 | NAGPA | 51172 | RNAseq | 0.2023 | 0.3839 | |
| SRP193095 | NAGPA | 51172 | RNAseq | -0.1446 | 0.5140 | |
| SRP219564 | NAGPA | 51172 | RNAseq | 0.8594 | 0.0508 | |
| TCGA | NAGPA | 51172 | RNAseq | -0.0787 | 0.2046 |
Upregulated datasets: 0; Downregulated datasets: 0.
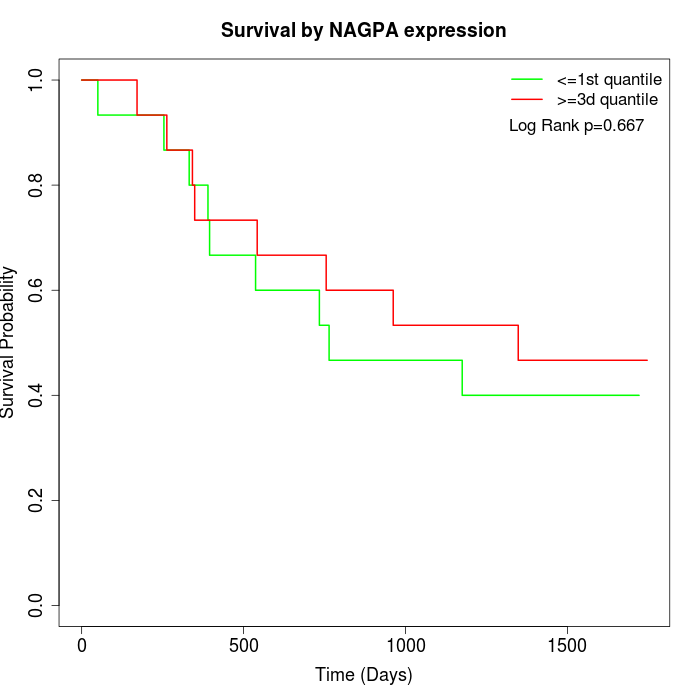
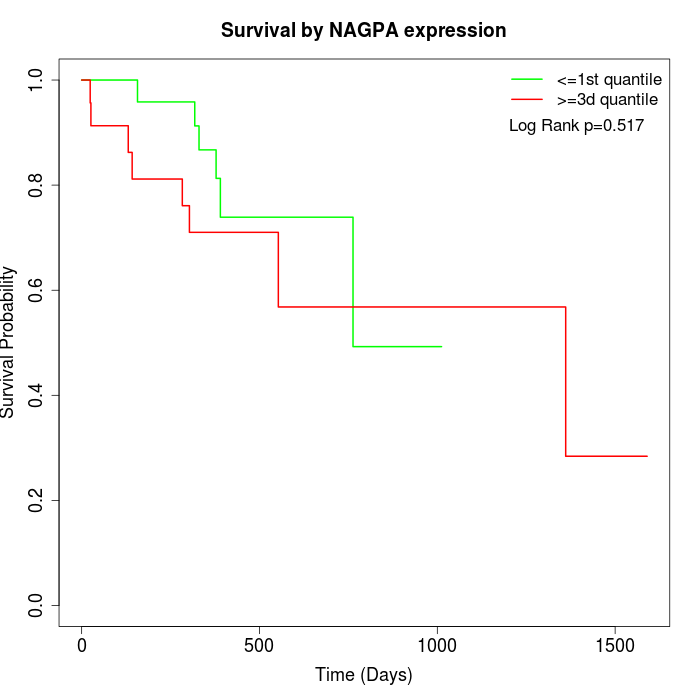
Survival by NAGPA expression:
Note: Click image to view full size file.
Copy number change of NAGPA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAGPA | 51172 | 5 | 4 | 21 | |
| GSE20123 | NAGPA | 51172 | 6 | 3 | 21 | |
| GSE43470 | NAGPA | 51172 | 4 | 4 | 35 | |
| GSE46452 | NAGPA | 51172 | 38 | 1 | 20 | |
| GSE47630 | NAGPA | 51172 | 13 | 6 | 21 | |
| GSE54993 | NAGPA | 51172 | 3 | 5 | 62 | |
| GSE54994 | NAGPA | 51172 | 5 | 9 | 39 | |
| GSE60625 | NAGPA | 51172 | 4 | 0 | 7 | |
| GSE74703 | NAGPA | 51172 | 4 | 4 | 28 | |
| GSE74704 | NAGPA | 51172 | 3 | 2 | 15 | |
| TCGA | NAGPA | 51172 | 19 | 13 | 64 |
Total number of gains: 104; Total number of losses: 51; Total Number of normals: 333.
Somatic mutations of NAGPA:
Generating mutation plots.
Highly correlated genes for NAGPA:
Showing top 20/614 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAGPA | SLC39A10 | 0.709149 | 3 | 0 | 3 |
| NAGPA | SPNS1 | 0.701663 | 5 | 0 | 5 |
| NAGPA | MAD2L2 | 0.69243 | 5 | 0 | 4 |
| NAGPA | C16orf91 | 0.690616 | 3 | 0 | 3 |
| NAGPA | DCK | 0.681706 | 3 | 0 | 3 |
| NAGPA | TDRD5 | 0.676403 | 3 | 0 | 3 |
| NAGPA | TRAF7 | 0.675571 | 5 | 0 | 5 |
| NAGPA | NARF | 0.670838 | 3 | 0 | 3 |
| NAGPA | SWT1 | 0.670078 | 3 | 0 | 3 |
| NAGPA | SLC19A1 | 0.66906 | 3 | 0 | 3 |
| NAGPA | ZAP70 | 0.660699 | 3 | 0 | 3 |
| NAGPA | C1orf50 | 0.658724 | 3 | 0 | 3 |
| NAGPA | ZNF75A | 0.658596 | 3 | 0 | 3 |
| NAGPA | TRADD | 0.658445 | 3 | 0 | 3 |
| NAGPA | TRIM59 | 0.655486 | 3 | 0 | 3 |
| NAGPA | PDIA5 | 0.650495 | 3 | 0 | 3 |
| NAGPA | HDDC3 | 0.648797 | 3 | 0 | 3 |
| NAGPA | ZNF213 | 0.644558 | 3 | 0 | 3 |
| NAGPA | ALG1 | 0.644542 | 6 | 0 | 4 |
| NAGPA | C1orf74 | 0.642207 | 3 | 0 | 3 |
For details and further investigation, click here