| Full name: zeta chain of T cell receptor associated protein kinase 70 | Alias Symbol: ZAP-70|STD | ||
| Type: protein-coding gene | Cytoband: 2q11.2 | ||
| Entrez ID: 7535 | HGNC ID: HGNC:12858 | Ensembl Gene: ENSG00000115085 | OMIM ID: 176947 |
| Drug and gene relationship at DGIdb | |||
ZAP70 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway | |
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa04660 | T cell receptor signaling pathway |
Expression of ZAP70:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZAP70 | 7535 | 214032_at | 0.8206 | 0.2906 | |
| GSE20347 | ZAP70 | 7535 | 214032_at | 0.0466 | 0.7943 | |
| GSE23400 | ZAP70 | 7535 | 214032_at | -0.0499 | 0.3517 | |
| GSE26886 | ZAP70 | 7535 | 214032_at | 0.4522 | 0.0117 | |
| GSE29001 | ZAP70 | 7535 | 214032_at | 0.2085 | 0.2073 | |
| GSE38129 | ZAP70 | 7535 | 214032_at | -0.1394 | 0.4065 | |
| GSE45670 | ZAP70 | 7535 | 214032_at | 0.1186 | 0.3788 | |
| GSE53622 | ZAP70 | 7535 | 4580 | -0.0918 | 0.5581 | |
| GSE53624 | ZAP70 | 7535 | 4580 | -0.4274 | 0.0000 | |
| GSE63941 | ZAP70 | 7535 | 214032_at | -0.0194 | 0.9383 | |
| GSE77861 | ZAP70 | 7535 | 214032_at | 0.0229 | 0.8483 | |
| GSE97050 | ZAP70 | 7535 | A_24_P169234 | 0.5767 | 0.2521 | |
| SRP064894 | ZAP70 | 7535 | RNAseq | -0.2109 | 0.5526 | |
| SRP133303 | ZAP70 | 7535 | RNAseq | -0.8544 | 0.0095 | |
| SRP159526 | ZAP70 | 7535 | RNAseq | -1.4105 | 0.0541 | |
| SRP193095 | ZAP70 | 7535 | RNAseq | -0.6871 | 0.0130 | |
| SRP219564 | ZAP70 | 7535 | RNAseq | 0.1232 | 0.8162 | |
| TCGA | ZAP70 | 7535 | RNAseq | -0.2647 | 0.3469 |
Upregulated datasets: 0; Downregulated datasets: 0.
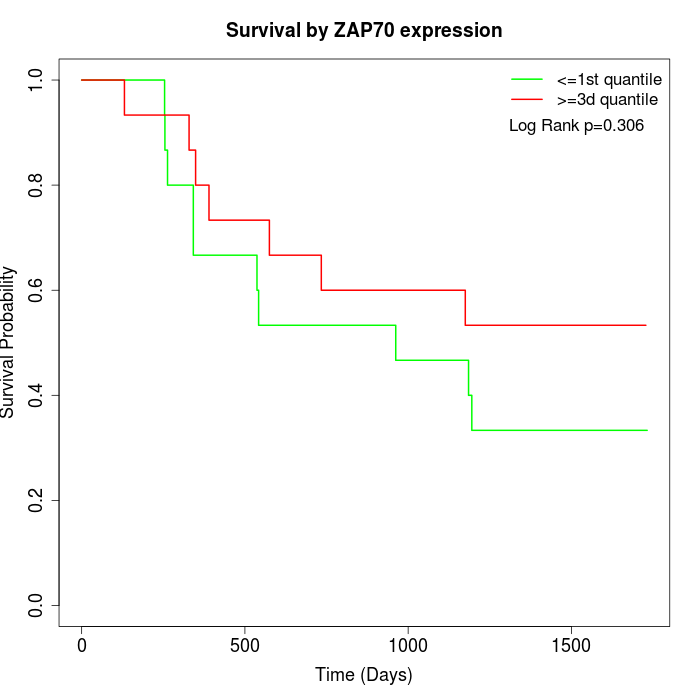
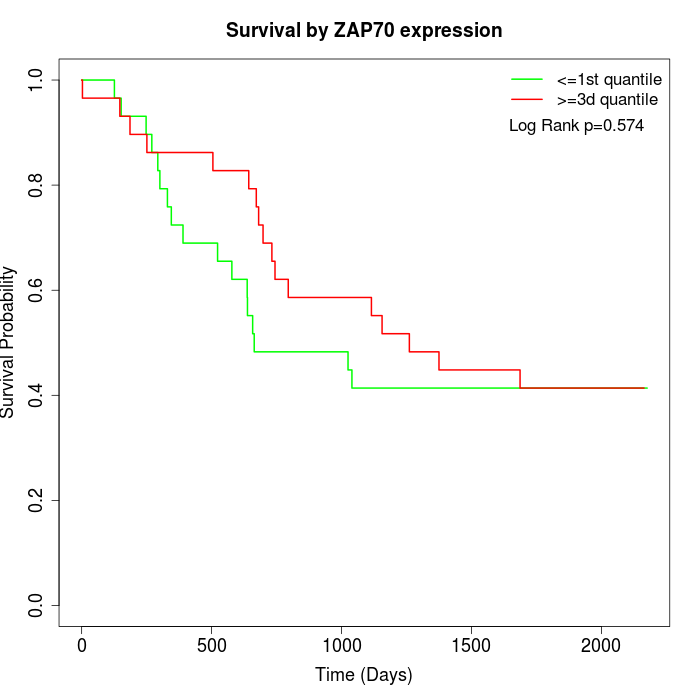
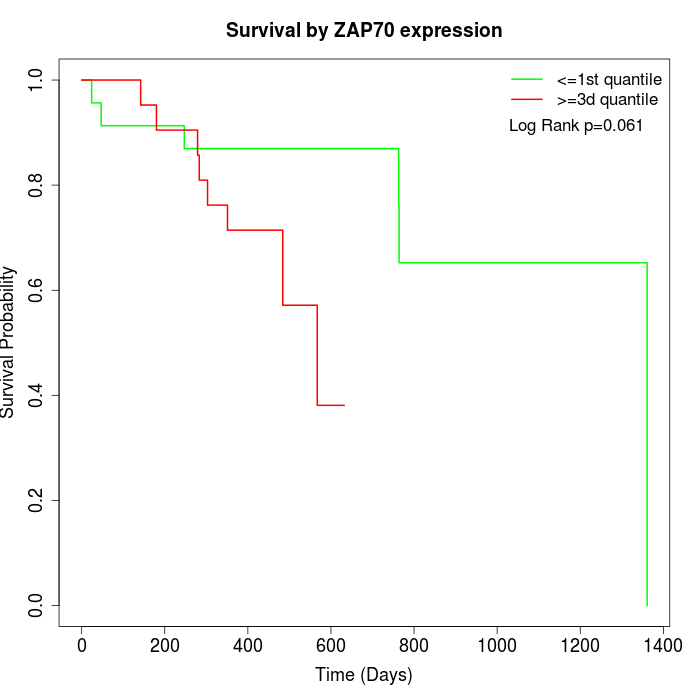
Survival by ZAP70 expression:
Note: Click image to view full size file.
Copy number change of ZAP70:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZAP70 | 7535 | 6 | 2 | 22 | |
| GSE20123 | ZAP70 | 7535 | 6 | 2 | 22 | |
| GSE43470 | ZAP70 | 7535 | 3 | 3 | 37 | |
| GSE46452 | ZAP70 | 7535 | 2 | 3 | 54 | |
| GSE47630 | ZAP70 | 7535 | 7 | 0 | 33 | |
| GSE54993 | ZAP70 | 7535 | 0 | 6 | 64 | |
| GSE54994 | ZAP70 | 7535 | 11 | 0 | 42 | |
| GSE60625 | ZAP70 | 7535 | 0 | 3 | 8 | |
| GSE74703 | ZAP70 | 7535 | 3 | 2 | 31 | |
| GSE74704 | ZAP70 | 7535 | 4 | 1 | 15 | |
| TCGA | ZAP70 | 7535 | 35 | 3 | 58 |
Total number of gains: 77; Total number of losses: 25; Total Number of normals: 386.
Somatic mutations of ZAP70:
Generating mutation plots.
Highly correlated genes for ZAP70:
Showing top 20/665 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZAP70 | TIMM8A | 0.793406 | 3 | 0 | 3 |
| ZAP70 | PREX1 | 0.783969 | 3 | 0 | 3 |
| ZAP70 | TNS4 | 0.774973 | 3 | 0 | 3 |
| ZAP70 | SNRPD2 | 0.758356 | 3 | 0 | 3 |
| ZAP70 | CNPY2 | 0.756114 | 3 | 0 | 3 |
| ZAP70 | TIGIT | 0.749532 | 6 | 0 | 6 |
| ZAP70 | ACAP1 | 0.749048 | 7 | 0 | 7 |
| ZAP70 | RCN1 | 0.746577 | 3 | 0 | 3 |
| ZAP70 | APOE | 0.744673 | 3 | 0 | 3 |
| ZAP70 | PLA2G7 | 0.742941 | 3 | 0 | 3 |
| ZAP70 | ZNF473 | 0.739992 | 3 | 0 | 3 |
| ZAP70 | HS6ST1 | 0.73941 | 3 | 0 | 3 |
| ZAP70 | MARCKSL1 | 0.738803 | 4 | 0 | 4 |
| ZAP70 | CASP8 | 0.735284 | 3 | 0 | 3 |
| ZAP70 | SUSD3 | 0.732437 | 5 | 0 | 5 |
| ZAP70 | SSBP4 | 0.732041 | 3 | 0 | 3 |
| ZAP70 | LAMP3 | 0.73165 | 4 | 0 | 4 |
| ZAP70 | POU2AF1 | 0.730678 | 3 | 0 | 3 |
| ZAP70 | DTL | 0.729358 | 3 | 0 | 3 |
| ZAP70 | EFTUD2 | 0.728129 | 3 | 0 | 3 |
For details and further investigation, click here