| Full name: nuclear transcription factor Y subunit alpha | Alias Symbol: HAP2|CBF-B|NF-YA | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 4800 | HGNC ID: HGNC:7804 | Ensembl Gene: ENSG00000001167 | OMIM ID: 189903 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NFYA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation | |
| hsa05152 | Tuberculosis |
Expression of NFYA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NFYA | 4800 | 204108_at | 1.1631 | 0.0647 | |
| GSE20347 | NFYA | 4800 | 204108_at | 0.6115 | 0.0006 | |
| GSE23400 | NFYA | 4800 | 204108_at | 0.4725 | 0.0000 | |
| GSE26886 | NFYA | 4800 | 204108_at | -0.5968 | 0.0410 | |
| GSE29001 | NFYA | 4800 | 204108_at | 0.5575 | 0.0701 | |
| GSE38129 | NFYA | 4800 | 204108_at | 0.6414 | 0.0000 | |
| GSE45670 | NFYA | 4800 | 204108_at | 0.7417 | 0.0000 | |
| GSE53622 | NFYA | 4800 | 12477 | 0.5855 | 0.0000 | |
| GSE53624 | NFYA | 4800 | 12477 | 0.8847 | 0.0000 | |
| GSE63941 | NFYA | 4800 | 204108_at | -0.5304 | 0.0890 | |
| GSE77861 | NFYA | 4800 | 215720_s_at | -0.0486 | 0.7819 | |
| GSE97050 | NFYA | 4800 | A_24_P414719 | -0.2798 | 0.4683 | |
| SRP007169 | NFYA | 4800 | RNAseq | -0.0264 | 0.9436 | |
| SRP008496 | NFYA | 4800 | RNAseq | 0.2887 | 0.2774 | |
| SRP064894 | NFYA | 4800 | RNAseq | 0.4541 | 0.0453 | |
| SRP133303 | NFYA | 4800 | RNAseq | 0.4612 | 0.0015 | |
| SRP159526 | NFYA | 4800 | RNAseq | 0.3190 | 0.1491 | |
| SRP193095 | NFYA | 4800 | RNAseq | 0.0764 | 0.4850 | |
| SRP219564 | NFYA | 4800 | RNAseq | 0.3834 | 0.3036 | |
| TCGA | NFYA | 4800 | RNAseq | -0.0068 | 0.8948 |
Upregulated datasets: 0; Downregulated datasets: 0.
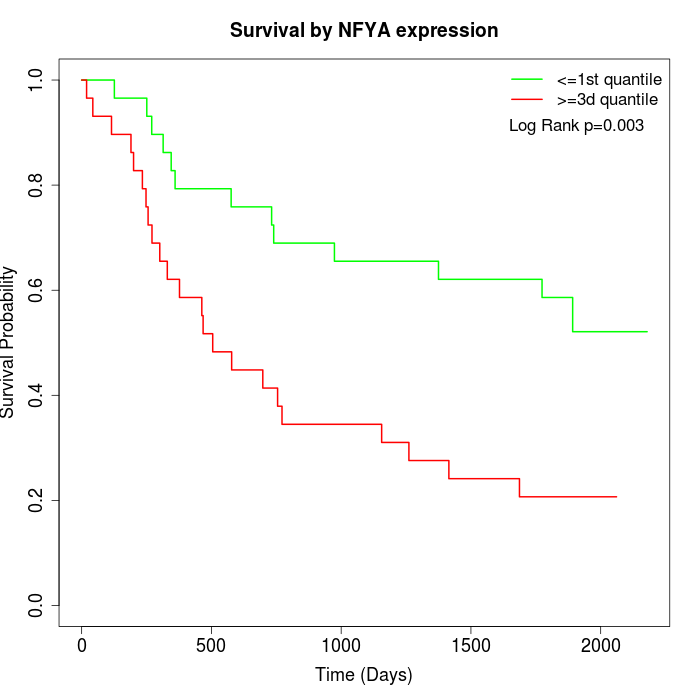
Survival by NFYA expression:
Note: Click image to view full size file.
Copy number change of NFYA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NFYA | 4800 | 3 | 3 | 24 | |
| GSE20123 | NFYA | 4800 | 3 | 3 | 24 | |
| GSE43470 | NFYA | 4800 | 6 | 0 | 37 | |
| GSE46452 | NFYA | 4800 | 2 | 9 | 48 | |
| GSE47630 | NFYA | 4800 | 8 | 4 | 28 | |
| GSE54993 | NFYA | 4800 | 3 | 2 | 65 | |
| GSE54994 | NFYA | 4800 | 11 | 4 | 38 | |
| GSE60625 | NFYA | 4800 | 0 | 1 | 10 | |
| GSE74703 | NFYA | 4800 | 6 | 0 | 30 | |
| GSE74704 | NFYA | 4800 | 1 | 3 | 16 | |
| TCGA | NFYA | 4800 | 19 | 13 | 64 |
Total number of gains: 62; Total number of losses: 42; Total Number of normals: 384.
Somatic mutations of NFYA:
Generating mutation plots.
Highly correlated genes for NFYA:
Showing top 20/158 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NFYA | LAT | 0.773194 | 3 | 0 | 3 |
| NFYA | ANKLE1 | 0.718457 | 3 | 0 | 3 |
| NFYA | ZNF444 | 0.711469 | 3 | 0 | 3 |
| NFYA | GPC2 | 0.68101 | 4 | 0 | 4 |
| NFYA | RAD54B | 0.672548 | 9 | 0 | 8 |
| NFYA | ECT2 | 0.669187 | 7 | 0 | 7 |
| NFYA | CDCA5 | 0.658471 | 5 | 0 | 5 |
| NFYA | GPR180 | 0.656871 | 5 | 0 | 3 |
| NFYA | MED20 | 0.656396 | 10 | 0 | 8 |
| NFYA | MTERF3 | 0.652121 | 6 | 0 | 5 |
| NFYA | TUBB | 0.647935 | 8 | 0 | 8 |
| NFYA | MEX3D | 0.646018 | 6 | 0 | 5 |
| NFYA | SERPINH1 | 0.644305 | 8 | 0 | 7 |
| NFYA | SCN9A | 0.64374 | 3 | 0 | 3 |
| NFYA | ING5 | 0.642322 | 4 | 0 | 3 |
| NFYA | CKS2 | 0.640903 | 7 | 0 | 6 |
| NFYA | SPDL1 | 0.639743 | 7 | 0 | 7 |
| NFYA | BAX | 0.633587 | 6 | 0 | 5 |
| NFYA | MEST | 0.631749 | 8 | 0 | 7 |
| NFYA | ZDHHC8 | 0.63128 | 5 | 0 | 3 |
For details and further investigation, click here