| Full name: linker for activation of T cells | Alias Symbol: LAT1 | ||
| Type: protein-coding gene | Cytoband: 16q13 | ||
| Entrez ID: 27040 | HGNC ID: HGNC:18874 | Ensembl Gene: ENSG00000213658 | OMIM ID: 602354 |
| Drug and gene relationship at DGIdb | |||
LAT involved pathways:
Expression of LAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAT | 27040 | 209881_s_at | 0.1706 | 0.5703 | |
| GSE20347 | LAT | 27040 | 209881_s_at | 0.1685 | 0.1393 | |
| GSE23400 | LAT | 27040 | 209881_s_at | -0.0500 | 0.2182 | |
| GSE26886 | LAT | 27040 | 209881_s_at | 0.4923 | 0.0108 | |
| GSE29001 | LAT | 27040 | 211005_at | 0.1891 | 0.7176 | |
| GSE38129 | LAT | 27040 | 209881_s_at | 0.0604 | 0.5530 | |
| GSE45670 | LAT | 27040 | 209881_s_at | 0.0995 | 0.4071 | |
| GSE63941 | LAT | 27040 | 209881_s_at | -0.1731 | 0.4574 | |
| GSE77861 | LAT | 27040 | 211005_at | -0.1691 | 0.1545 | |
| GSE97050 | LAT | 27040 | A_23_P44112 | -0.2770 | 0.3366 | |
| SRP064894 | LAT | 27040 | RNAseq | 0.2595 | 0.3835 | |
| SRP133303 | LAT | 27040 | RNAseq | -0.5179 | 0.0157 | |
| SRP159526 | LAT | 27040 | RNAseq | -0.4325 | 0.1765 | |
| SRP193095 | LAT | 27040 | RNAseq | 0.0421 | 0.8399 | |
| SRP219564 | LAT | 27040 | RNAseq | -0.0234 | 0.9578 | |
| TCGA | LAT | 27040 | RNAseq | 0.0682 | 0.6507 |
Upregulated datasets: 0; Downregulated datasets: 0.
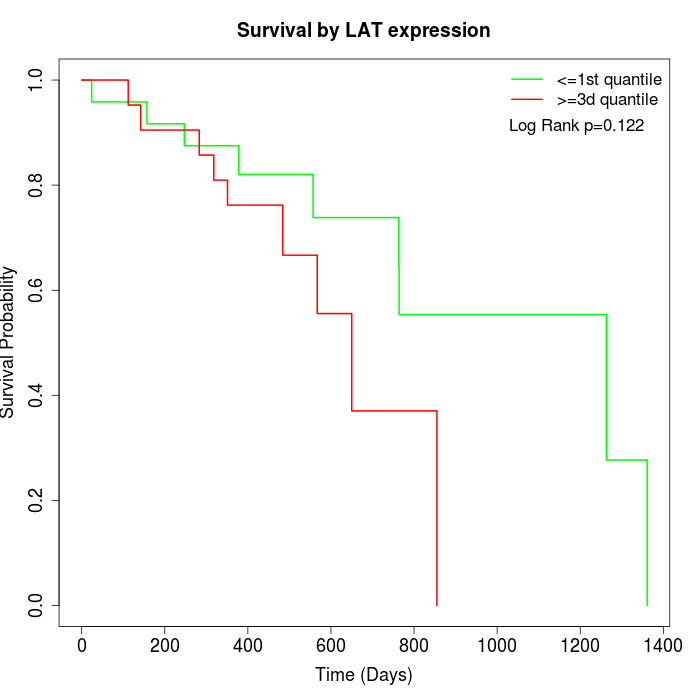
Survival by LAT expression:
Note: Click image to view full size file.
Copy number change of LAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAT | 27040 | 5 | 5 | 20 | |
| GSE20123 | LAT | 27040 | 5 | 4 | 21 | |
| GSE43470 | LAT | 27040 | 3 | 3 | 37 | |
| GSE46452 | LAT | 27040 | 38 | 1 | 20 | |
| GSE47630 | LAT | 27040 | 11 | 7 | 22 | |
| GSE54993 | LAT | 27040 | 3 | 5 | 62 | |
| GSE54994 | LAT | 27040 | 5 | 9 | 39 | |
| GSE60625 | LAT | 27040 | 4 | 0 | 7 | |
| GSE74703 | LAT | 27040 | 3 | 2 | 31 | |
| GSE74704 | LAT | 27040 | 3 | 2 | 15 | |
| TCGA | LAT | 27040 | 20 | 12 | 64 |
Total number of gains: 100; Total number of losses: 50; Total Number of normals: 338.
Somatic mutations of LAT:
Generating mutation plots.
Highly correlated genes for LAT:
Showing top 20/324 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAT | NFYA | 0.773194 | 3 | 0 | 3 |
| LAT | B3GAT3 | 0.743837 | 3 | 0 | 3 |
| LAT | PNPLA7 | 0.742696 | 3 | 0 | 3 |
| LAT | LTBP3 | 0.737664 | 3 | 0 | 3 |
| LAT | BRSK1 | 0.721215 | 3 | 0 | 3 |
| LAT | CPAMD8 | 0.720477 | 3 | 0 | 3 |
| LAT | PRRT2 | 0.716379 | 3 | 0 | 3 |
| LAT | MEX3D | 0.713499 | 3 | 0 | 3 |
| LAT | IL17B | 0.711123 | 4 | 0 | 4 |
| LAT | CATSPER3 | 0.696488 | 3 | 0 | 3 |
| LAT | ZNF341 | 0.695818 | 4 | 0 | 3 |
| LAT | TXNDC2 | 0.693821 | 3 | 0 | 3 |
| LAT | SP8 | 0.693515 | 3 | 0 | 3 |
| LAT | PPP2R2B | 0.692326 | 3 | 0 | 3 |
| LAT | TNN | 0.692073 | 5 | 0 | 4 |
| LAT | PCED1A | 0.691826 | 3 | 0 | 3 |
| LAT | ABCA2 | 0.690724 | 5 | 0 | 4 |
| LAT | ADAMTS2 | 0.68781 | 3 | 0 | 3 |
| LAT | SCARF2 | 0.683385 | 4 | 0 | 3 |
| LAT | SYCE1L | 0.678109 | 5 | 0 | 4 |
For details and further investigation, click here