| Full name: epithelial cell transforming 2 | Alias Symbol: ARHGEF31 | ||
| Type: protein-coding gene | Cytoband: 3q26.31 | ||
| Entrez ID: 1894 | HGNC ID: HGNC:3155 | Ensembl Gene: ENSG00000114346 | OMIM ID: 600586 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ECT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ECT2 | 1894 | 219787_s_at | 1.9498 | 0.0132 | |
| GSE20347 | ECT2 | 1894 | 219787_s_at | 2.5337 | 0.0000 | |
| GSE23400 | ECT2 | 1894 | 219787_s_at | 2.3146 | 0.0000 | |
| GSE26886 | ECT2 | 1894 | 219787_s_at | 1.1950 | 0.0024 | |
| GSE29001 | ECT2 | 1894 | 219787_s_at | 2.3546 | 0.0001 | |
| GSE38129 | ECT2 | 1894 | 219787_s_at | 2.6888 | 0.0000 | |
| GSE45670 | ECT2 | 1894 | 219787_s_at | 1.8485 | 0.0000 | |
| GSE53622 | ECT2 | 1894 | 57984 | 1.8340 | 0.0000 | |
| GSE53624 | ECT2 | 1894 | 57984 | 2.1332 | 0.0000 | |
| GSE63941 | ECT2 | 1894 | 219787_s_at | 1.5904 | 0.0057 | |
| GSE77861 | ECT2 | 1894 | 219787_s_at | 1.7290 | 0.0042 | |
| GSE97050 | ECT2 | 1894 | A_23_P44684 | 0.7755 | 0.1235 | |
| SRP007169 | ECT2 | 1894 | RNAseq | 2.9767 | 0.0000 | |
| SRP008496 | ECT2 | 1894 | RNAseq | 2.9551 | 0.0000 | |
| SRP064894 | ECT2 | 1894 | RNAseq | 1.9029 | 0.0000 | |
| SRP133303 | ECT2 | 1894 | RNAseq | 2.2374 | 0.0000 | |
| SRP159526 | ECT2 | 1894 | RNAseq | 1.9618 | 0.0000 | |
| SRP193095 | ECT2 | 1894 | RNAseq | 1.6423 | 0.0000 | |
| SRP219564 | ECT2 | 1894 | RNAseq | 1.5602 | 0.0166 | |
| TCGA | ECT2 | 1894 | RNAseq | 0.7454 | 0.0000 |
Upregulated datasets: 18; Downregulated datasets: 0.
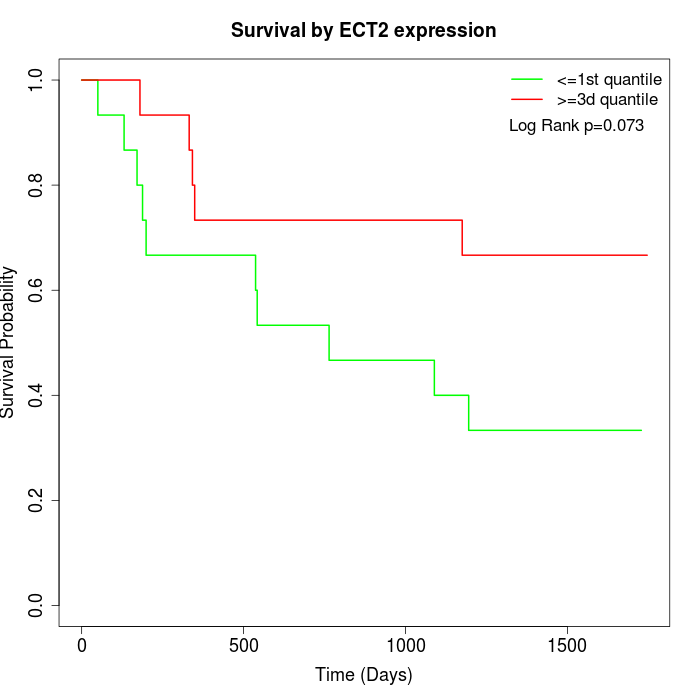
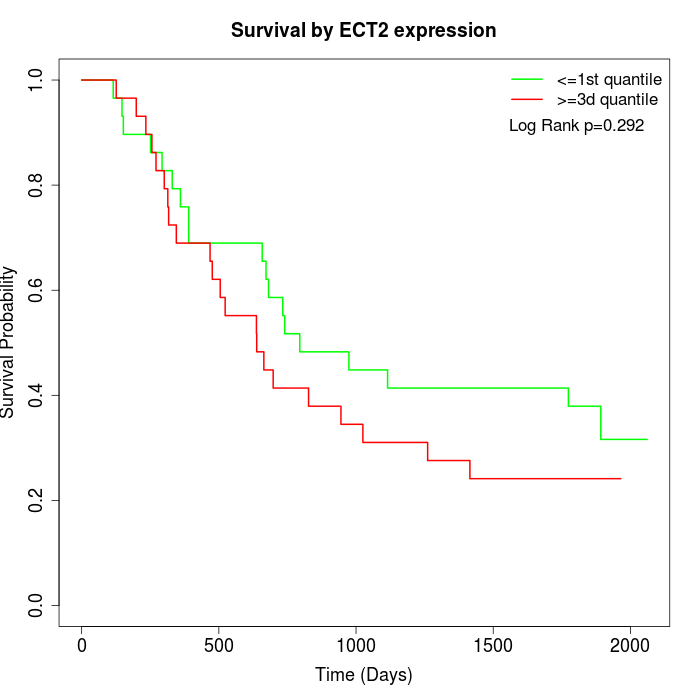
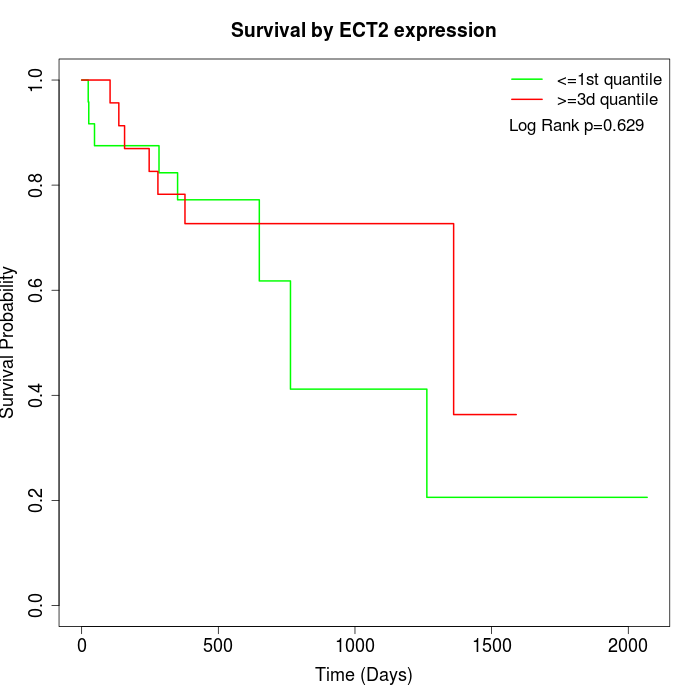
Survival by ECT2 expression:
Note: Click image to view full size file.
Copy number change of ECT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ECT2 | 1894 | 24 | 0 | 6 | |
| GSE20123 | ECT2 | 1894 | 24 | 0 | 6 | |
| GSE43470 | ECT2 | 1894 | 23 | 0 | 20 | |
| GSE46452 | ECT2 | 1894 | 19 | 1 | 39 | |
| GSE47630 | ECT2 | 1894 | 27 | 2 | 11 | |
| GSE54993 | ECT2 | 1894 | 1 | 15 | 54 | |
| GSE54994 | ECT2 | 1894 | 41 | 0 | 12 | |
| GSE60625 | ECT2 | 1894 | 0 | 6 | 5 | |
| GSE74703 | ECT2 | 1894 | 19 | 0 | 17 | |
| GSE74704 | ECT2 | 1894 | 16 | 0 | 4 | |
| TCGA | ECT2 | 1894 | 79 | 0 | 17 |
Total number of gains: 273; Total number of losses: 24; Total Number of normals: 191.
Somatic mutations of ECT2:
Generating mutation plots.
Highly correlated genes for ECT2:
Showing top 20/2005 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ECT2 | CDK1 | 0.869633 | 12 | 0 | 12 |
| ECT2 | RAD51AP1 | 0.858846 | 13 | 0 | 13 |
| ECT2 | ACTL6A | 0.847076 | 12 | 0 | 12 |
| ECT2 | CKS1B | 0.846941 | 12 | 0 | 12 |
| ECT2 | RFC4 | 0.846739 | 13 | 0 | 13 |
| ECT2 | TRIP13 | 0.834287 | 12 | 0 | 12 |
| ECT2 | CBX3 | 0.833697 | 11 | 0 | 11 |
| ECT2 | TPX2 | 0.832086 | 12 | 0 | 12 |
| ECT2 | KIF4A | 0.827784 | 12 | 0 | 11 |
| ECT2 | AURKB | 0.826706 | 12 | 0 | 12 |
| ECT2 | NEK2 | 0.821662 | 13 | 0 | 12 |
| ECT2 | KIF14 | 0.821485 | 13 | 0 | 12 |
| ECT2 | FOXM1 | 0.817701 | 13 | 0 | 12 |
| ECT2 | MCM6 | 0.817198 | 12 | 0 | 12 |
| ECT2 | GMPS | 0.81547 | 11 | 0 | 11 |
| ECT2 | CEP55 | 0.814792 | 13 | 0 | 12 |
| ECT2 | TIMELESS | 0.814533 | 12 | 0 | 12 |
| ECT2 | CENPF | 0.813821 | 13 | 0 | 13 |
| ECT2 | TOP2A | 0.813161 | 13 | 0 | 13 |
| ECT2 | ATAD2 | 0.812511 | 12 | 0 | 11 |
For details and further investigation, click here