| Full name: nitrilase family member 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q12.2 | ||
| Entrez ID: 56954 | HGNC ID: HGNC:29878 | Ensembl Gene: ENSG00000114021 | OMIM ID: 616769 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of NIT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIT2 | 56954 | 218557_at | 0.4612 | 0.2245 | |
| GSE20347 | NIT2 | 56954 | 218557_at | 0.7544 | 0.0002 | |
| GSE23400 | NIT2 | 56954 | 218557_at | 0.7376 | 0.0000 | |
| GSE26886 | NIT2 | 56954 | 218557_at | -0.3599 | 0.1575 | |
| GSE29001 | NIT2 | 56954 | 218557_at | 0.4150 | 0.4309 | |
| GSE38129 | NIT2 | 56954 | 218557_at | 0.8728 | 0.0000 | |
| GSE45670 | NIT2 | 56954 | 218557_at | 0.6480 | 0.0001 | |
| GSE53622 | NIT2 | 56954 | 19551 | 0.4207 | 0.0000 | |
| GSE53624 | NIT2 | 56954 | 19551 | 0.6557 | 0.0000 | |
| GSE63941 | NIT2 | 56954 | 218557_at | 0.6245 | 0.1971 | |
| GSE77861 | NIT2 | 56954 | 218557_at | 0.2590 | 0.5940 | |
| GSE97050 | NIT2 | 56954 | A_24_P120346 | 0.2433 | 0.3485 | |
| SRP007169 | NIT2 | 56954 | RNAseq | -0.1755 | 0.6540 | |
| SRP008496 | NIT2 | 56954 | RNAseq | 0.2147 | 0.4380 | |
| SRP064894 | NIT2 | 56954 | RNAseq | 0.6257 | 0.0042 | |
| SRP133303 | NIT2 | 56954 | RNAseq | 0.0533 | 0.7751 | |
| SRP159526 | NIT2 | 56954 | RNAseq | 0.3013 | 0.2451 | |
| SRP193095 | NIT2 | 56954 | RNAseq | 0.1914 | 0.1009 | |
| SRP219564 | NIT2 | 56954 | RNAseq | 0.1705 | 0.4714 | |
| TCGA | NIT2 | 56954 | RNAseq | 0.2865 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
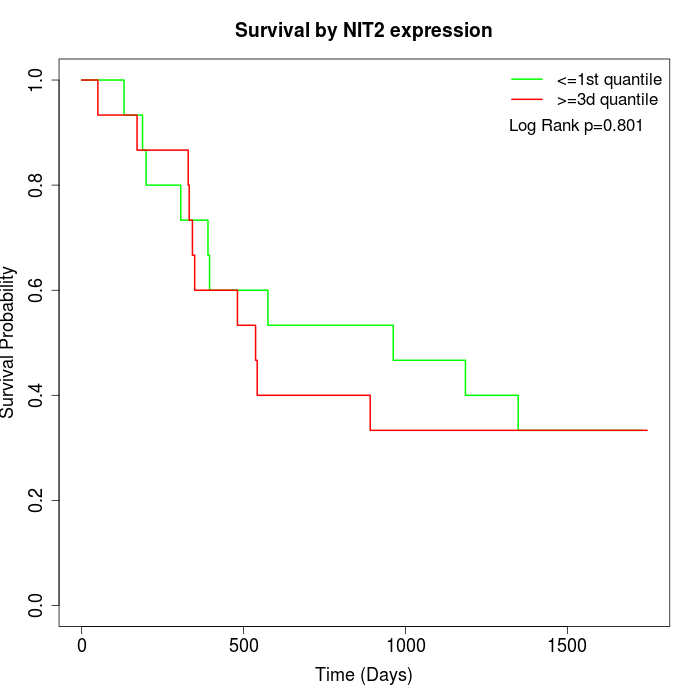
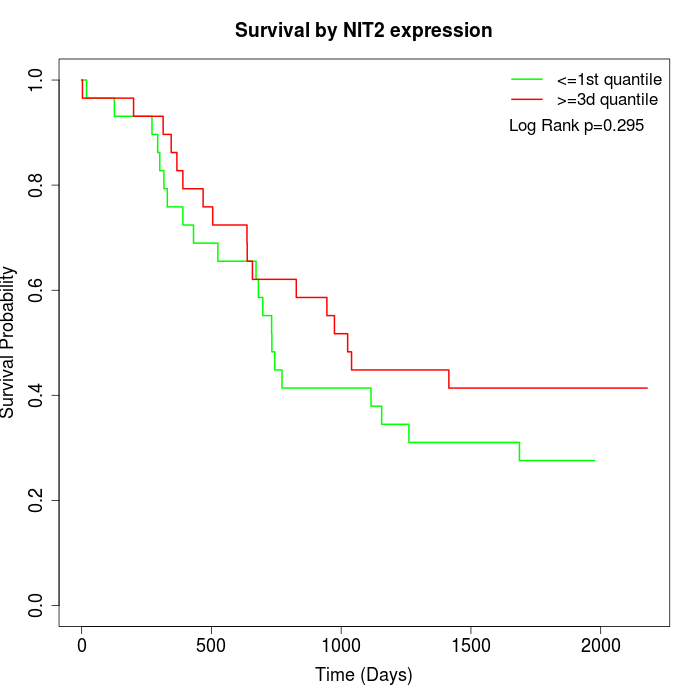
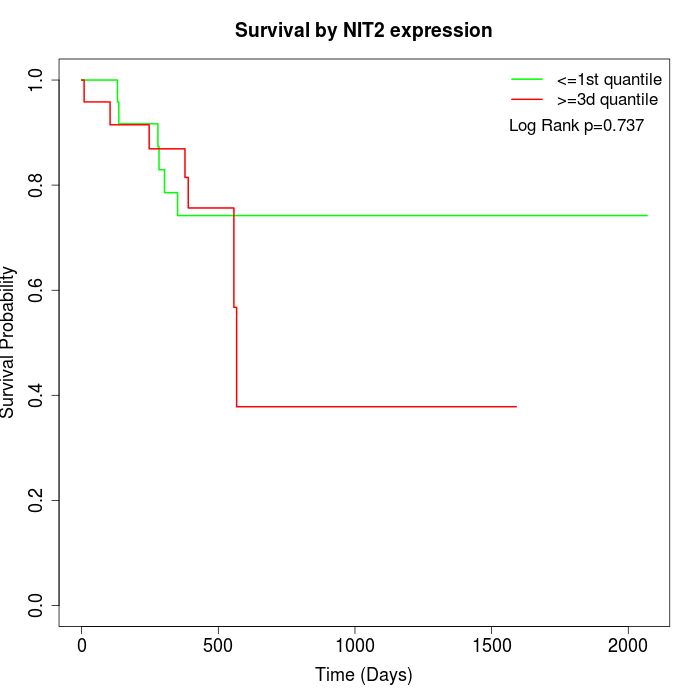
Survival by NIT2 expression:
Note: Click image to view full size file.
Copy number change of NIT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIT2 | 56954 | 17 | 1 | 12 | |
| GSE20123 | NIT2 | 56954 | 16 | 1 | 13 | |
| GSE43470 | NIT2 | 56954 | 19 | 1 | 23 | |
| GSE46452 | NIT2 | 56954 | 12 | 6 | 41 | |
| GSE47630 | NIT2 | 56954 | 16 | 6 | 18 | |
| GSE54993 | NIT2 | 56954 | 2 | 5 | 63 | |
| GSE54994 | NIT2 | 56954 | 27 | 5 | 21 | |
| GSE60625 | NIT2 | 56954 | 0 | 6 | 5 | |
| GSE74703 | NIT2 | 56954 | 15 | 1 | 20 | |
| GSE74704 | NIT2 | 56954 | 13 | 1 | 6 | |
| TCGA | NIT2 | 56954 | 48 | 10 | 38 |
Total number of gains: 185; Total number of losses: 43; Total Number of normals: 260.
Somatic mutations of NIT2:
Generating mutation plots.
Highly correlated genes for NIT2:
Showing top 20/1123 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIT2 | ZGPAT | 0.847068 | 3 | 0 | 3 |
| NIT2 | VPS28 | 0.784201 | 3 | 0 | 3 |
| NIT2 | FYTTD1 | 0.777067 | 3 | 0 | 3 |
| NIT2 | NFKB1 | 0.766105 | 3 | 0 | 3 |
| NIT2 | ATF7IP | 0.746792 | 3 | 0 | 3 |
| NIT2 | SYMPK | 0.74541 | 3 | 0 | 3 |
| NIT2 | KRTCAP2 | 0.744535 | 3 | 0 | 3 |
| NIT2 | GPX1 | 0.737087 | 3 | 0 | 3 |
| NIT2 | TRAF7 | 0.736171 | 3 | 0 | 3 |
| NIT2 | ISY1 | 0.734779 | 3 | 0 | 3 |
| NIT2 | METTL8 | 0.731438 | 5 | 0 | 4 |
| NIT2 | ISCA2 | 0.728156 | 3 | 0 | 3 |
| NIT2 | XRN1 | 0.723469 | 3 | 0 | 3 |
| NIT2 | DNAJC11 | 0.723404 | 3 | 0 | 3 |
| NIT2 | NLK | 0.721256 | 3 | 0 | 3 |
| NIT2 | TPP2 | 0.719717 | 3 | 0 | 3 |
| NIT2 | POLR2K | 0.716691 | 6 | 0 | 6 |
| NIT2 | COX16 | 0.715832 | 3 | 0 | 3 |
| NIT2 | TP53RK | 0.715217 | 4 | 0 | 3 |
| NIT2 | U2SURP | 0.712845 | 7 | 0 | 6 |
For details and further investigation, click here