| Full name: neuroligin 4 X-linked | Alias Symbol: KIAA1260|NLGN|HLNX | ||
| Type: protein-coding gene | Cytoband: Xp22.32-p22.31 | ||
| Entrez ID: 57502 | HGNC ID: HGNC:14287 | Ensembl Gene: ENSG00000146938 | OMIM ID: 300427 |
| Drug and gene relationship at DGIdb | |||
Expression of NLGN4X:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLGN4X | 57502 | 221933_at | 0.4243 | 0.7599 | |
| GSE20347 | NLGN4X | 57502 | 221933_at | 0.2192 | 0.2434 | |
| GSE23400 | NLGN4X | 57502 | 221933_at | 0.1840 | 0.0246 | |
| GSE26886 | NLGN4X | 57502 | 221933_at | 0.9721 | 0.0456 | |
| GSE29001 | NLGN4X | 57502 | 221933_at | 0.3789 | 0.5475 | |
| GSE38129 | NLGN4X | 57502 | 221933_at | -0.2470 | 0.4951 | |
| GSE45670 | NLGN4X | 57502 | 221933_at | 0.2176 | 0.7642 | |
| GSE53622 | NLGN4X | 57502 | 60500 | 0.5566 | 0.0015 | |
| GSE53624 | NLGN4X | 57502 | 60500 | 0.8600 | 0.0000 | |
| GSE63941 | NLGN4X | 57502 | 1554689_a_at | 0.0791 | 0.7980 | |
| GSE77861 | NLGN4X | 57502 | 1554689_a_at | -0.1741 | 0.0577 | |
| GSE97050 | NLGN4X | 57502 | A_33_P3287158 | 0.3614 | 0.5154 | |
| SRP133303 | NLGN4X | 57502 | RNAseq | 1.7481 | 0.0000 | |
| SRP159526 | NLGN4X | 57502 | RNAseq | 1.1596 | 0.0080 | |
| SRP219564 | NLGN4X | 57502 | RNAseq | 1.6256 | 0.0447 | |
| TCGA | NLGN4X | 57502 | RNAseq | 0.0543 | 0.8193 |
Upregulated datasets: 3; Downregulated datasets: 0.
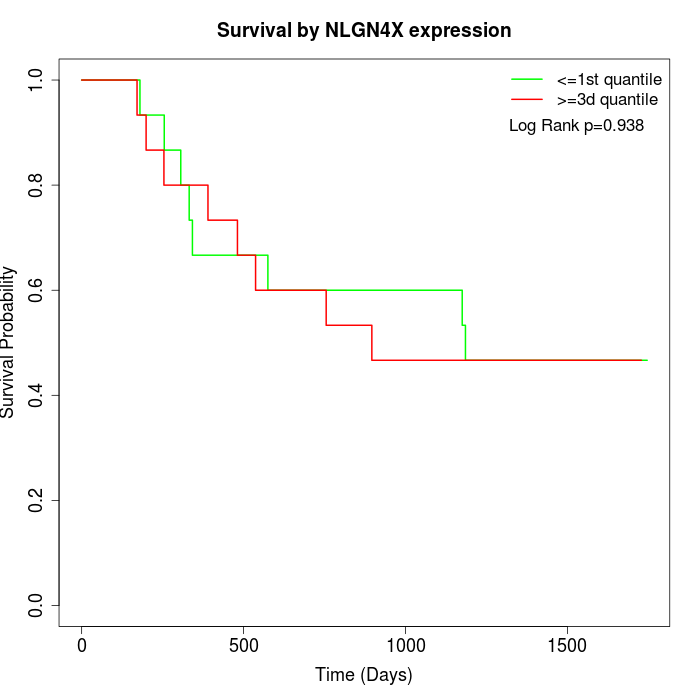
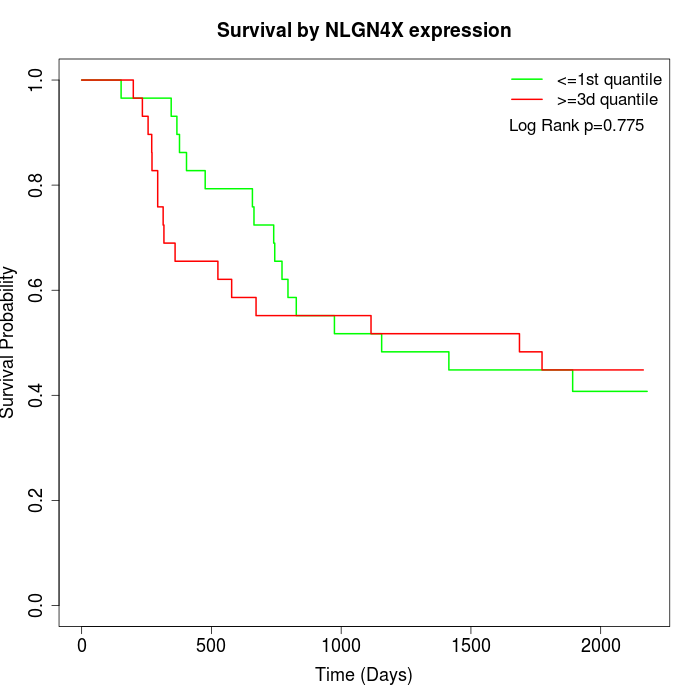
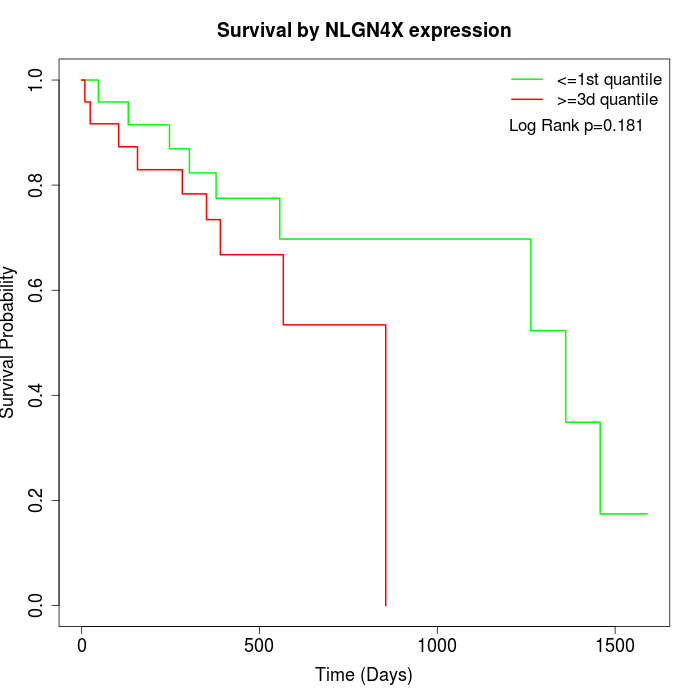
Survival by NLGN4X expression:
Note: Click image to view full size file.
Copy number change of NLGN4X:
No record found for this gene.
Somatic mutations of NLGN4X:
Generating mutation plots.
Highly correlated genes for NLGN4X:
Showing top 20/149 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLGN4X | KANK2 | 0.788915 | 3 | 0 | 3 |
| NLGN4X | TNP2 | 0.781597 | 3 | 0 | 3 |
| NLGN4X | ZDHHC1 | 0.781467 | 3 | 0 | 3 |
| NLGN4X | GRK5 | 0.777019 | 3 | 0 | 3 |
| NLGN4X | ARHGEF6 | 0.763061 | 3 | 0 | 3 |
| NLGN4X | FLI1 | 0.753208 | 3 | 0 | 3 |
| NLGN4X | FNBP1 | 0.742309 | 3 | 0 | 3 |
| NLGN4X | EPAS1 | 0.73819 | 3 | 0 | 3 |
| NLGN4X | STARD13 | 0.721166 | 3 | 0 | 3 |
| NLGN4X | RNASEH2C | 0.719752 | 3 | 0 | 3 |
| NLGN4X | SLIT3 | 0.71728 | 3 | 0 | 3 |
| NLGN4X | SOBP | 0.713607 | 3 | 0 | 3 |
| NLGN4X | USP36 | 0.712334 | 3 | 0 | 3 |
| NLGN4X | NMNAT2 | 0.707233 | 3 | 0 | 3 |
| NLGN4X | ZMYM6 | 0.704261 | 3 | 0 | 3 |
| NLGN4X | PLCE1 | 0.702171 | 3 | 0 | 3 |
| NLGN4X | IQSEC2 | 0.698635 | 3 | 0 | 3 |
| NLGN4X | NES | 0.697264 | 6 | 0 | 5 |
| NLGN4X | MYL9 | 0.693458 | 3 | 0 | 3 |
| NLGN4X | PDZRN4 | 0.692206 | 3 | 0 | 3 |
For details and further investigation, click here