| Full name: nucleotide binding oligomerization domain containing 1 | Alias Symbol: NLRC1|CLR7.1 | ||
| Type: protein-coding gene | Cytoband: 7p14.3 | ||
| Entrez ID: 10392 | HGNC ID: HGNC:16390 | Ensembl Gene: ENSG00000106100 | OMIM ID: 605980 |
| Drug and gene relationship at DGIdb | |||
NOD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05131 | Shigellosis | |
| hsa05133 | Pertussis |
Expression of NOD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NOD1 | 10392 | 221073_s_at | 0.0098 | 0.9733 | |
| GSE20347 | NOD1 | 10392 | 221073_s_at | -0.0279 | 0.7943 | |
| GSE23400 | NOD1 | 10392 | 221073_s_at | -0.0086 | 0.8386 | |
| GSE26886 | NOD1 | 10392 | 221073_s_at | 0.1443 | 0.3941 | |
| GSE29001 | NOD1 | 10392 | 221073_s_at | 0.0699 | 0.7262 | |
| GSE38129 | NOD1 | 10392 | 221073_s_at | -0.0597 | 0.5918 | |
| GSE45670 | NOD1 | 10392 | 221073_s_at | -0.1274 | 0.3373 | |
| GSE53622 | NOD1 | 10392 | 116233 | 0.1336 | 0.0951 | |
| GSE53624 | NOD1 | 10392 | 80684 | 0.3800 | 0.0000 | |
| GSE63941 | NOD1 | 10392 | 221073_s_at | 0.0975 | 0.7368 | |
| GSE77861 | NOD1 | 10392 | 221073_s_at | -0.0083 | 0.9609 | |
| GSE97050 | NOD1 | 10392 | A_24_P129277 | 0.4600 | 0.1296 | |
| SRP007169 | NOD1 | 10392 | RNAseq | 0.1572 | 0.7474 | |
| SRP008496 | NOD1 | 10392 | RNAseq | 0.5617 | 0.1878 | |
| SRP064894 | NOD1 | 10392 | RNAseq | 0.6366 | 0.0046 | |
| SRP133303 | NOD1 | 10392 | RNAseq | 0.2738 | 0.0957 | |
| SRP159526 | NOD1 | 10392 | RNAseq | 0.0347 | 0.8626 | |
| SRP193095 | NOD1 | 10392 | RNAseq | 0.4585 | 0.0001 | |
| SRP219564 | NOD1 | 10392 | RNAseq | 0.4195 | 0.1081 | |
| TCGA | NOD1 | 10392 | RNAseq | 0.1044 | 0.1396 |
Upregulated datasets: 0; Downregulated datasets: 0.
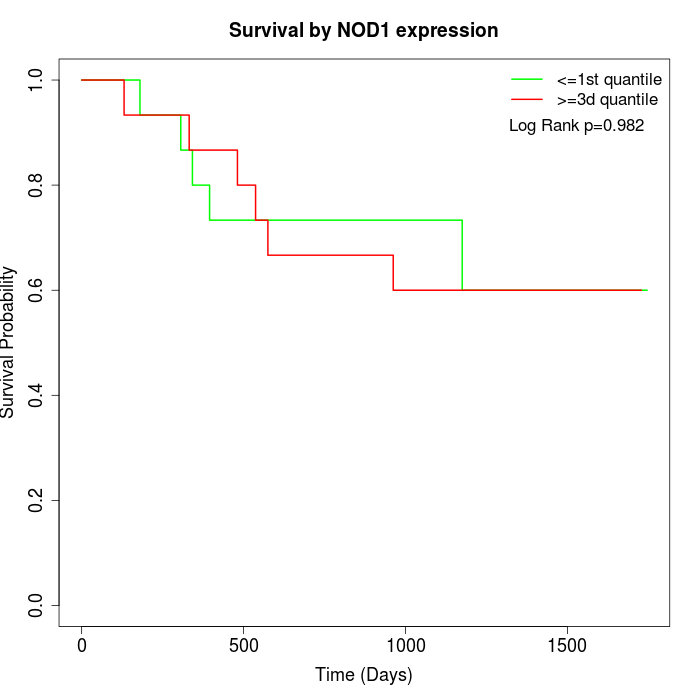
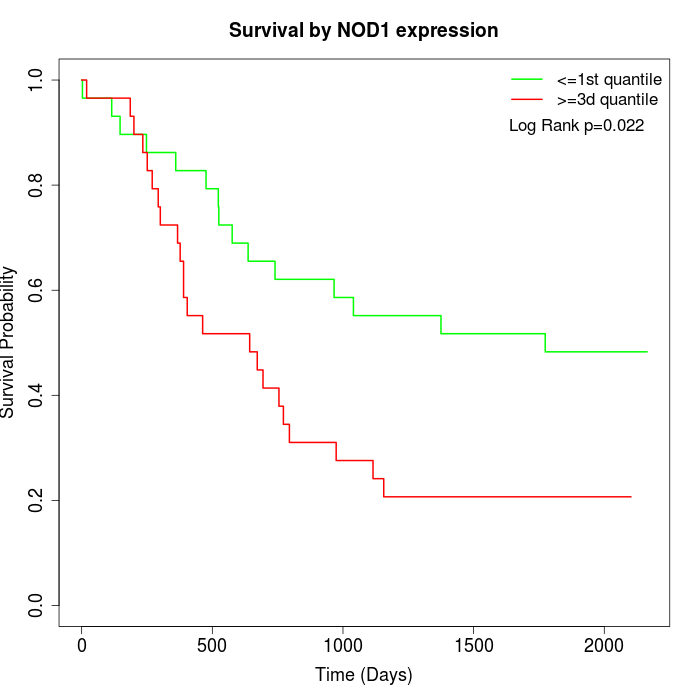
Survival by NOD1 expression:
Note: Click image to view full size file.
Copy number change of NOD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NOD1 | 10392 | 15 | 0 | 15 | |
| GSE20123 | NOD1 | 10392 | 15 | 0 | 15 | |
| GSE43470 | NOD1 | 10392 | 4 | 0 | 39 | |
| GSE46452 | NOD1 | 10392 | 11 | 1 | 47 | |
| GSE47630 | NOD1 | 10392 | 8 | 1 | 31 | |
| GSE54993 | NOD1 | 10392 | 0 | 6 | 64 | |
| GSE54994 | NOD1 | 10392 | 19 | 3 | 31 | |
| GSE60625 | NOD1 | 10392 | 0 | 0 | 11 | |
| GSE74703 | NOD1 | 10392 | 4 | 0 | 32 | |
| GSE74704 | NOD1 | 10392 | 9 | 0 | 11 | |
| TCGA | NOD1 | 10392 | 55 | 7 | 34 |
Total number of gains: 140; Total number of losses: 18; Total Number of normals: 330.
Somatic mutations of NOD1:
Generating mutation plots.
Highly correlated genes for NOD1:
Showing top 20/62 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NOD1 | ERVW-1 | 0.76398 | 3 | 0 | 3 |
| NOD1 | ACER3 | 0.749744 | 3 | 0 | 3 |
| NOD1 | STK36 | 0.705475 | 3 | 0 | 3 |
| NOD1 | ARCN1 | 0.684717 | 3 | 0 | 3 |
| NOD1 | ABHD1 | 0.6806 | 3 | 0 | 3 |
| NOD1 | SORD | 0.675638 | 3 | 0 | 3 |
| NOD1 | KCTD2 | 0.670271 | 3 | 0 | 3 |
| NOD1 | TLN1 | 0.669016 | 3 | 0 | 3 |
| NOD1 | LPCAT2 | 0.664575 | 4 | 0 | 3 |
| NOD1 | ADCY3 | 0.663521 | 3 | 0 | 3 |
| NOD1 | FGFR3 | 0.658894 | 3 | 0 | 3 |
| NOD1 | ITGB8 | 0.658558 | 3 | 0 | 3 |
| NOD1 | CCNYL1 | 0.655617 | 3 | 0 | 3 |
| NOD1 | TMEM17 | 0.654428 | 4 | 0 | 3 |
| NOD1 | TMEM41B | 0.653609 | 4 | 0 | 3 |
| NOD1 | FAM166A | 0.653375 | 3 | 0 | 3 |
| NOD1 | SLCO3A1 | 0.642672 | 4 | 0 | 3 |
| NOD1 | ELL3 | 0.631156 | 4 | 0 | 3 |
| NOD1 | GATAD2B | 0.630455 | 4 | 0 | 3 |
| NOD1 | BDKRB2 | 0.630261 | 4 | 0 | 3 |
For details and further investigation, click here