| Full name: nuclear receptor subfamily 1 group D member 2 | Alias Symbol: BD73|RVR|EAR-1r|HZF2|Hs.37288|REVERBB|REVERBbeta | ||
| Type: protein-coding gene | Cytoband: 3p24.2 | ||
| Entrez ID: 9975 | HGNC ID: HGNC:7963 | Ensembl Gene: ENSG00000174738 | OMIM ID: 602304 |
| Drug and gene relationship at DGIdb | |||
Expression of NR1D2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NR1D2 | 9975 | 225768_at | 0.1186 | 0.9230 | |
| GSE20347 | NR1D2 | 9975 | 209750_at | -0.3939 | 0.1591 | |
| GSE23400 | NR1D2 | 9975 | 209750_at | -0.4356 | 0.0000 | |
| GSE26886 | NR1D2 | 9975 | 225768_at | -1.7879 | 0.0000 | |
| GSE29001 | NR1D2 | 9975 | 209750_at | -0.7585 | 0.1954 | |
| GSE38129 | NR1D2 | 9975 | 209750_at | -0.4661 | 0.3258 | |
| GSE45670 | NR1D2 | 9975 | 225768_at | -0.5774 | 0.1454 | |
| GSE53622 | NR1D2 | 9975 | 2495 | -0.6329 | 0.0000 | |
| GSE53624 | NR1D2 | 9975 | 2495 | -0.5982 | 0.0000 | |
| GSE63941 | NR1D2 | 9975 | 209750_at | 1.0995 | 0.0959 | |
| GSE77861 | NR1D2 | 9975 | 225768_at | -1.0261 | 0.0699 | |
| GSE97050 | NR1D2 | 9975 | A_33_P3409124 | -0.0073 | 0.9864 | |
| SRP007169 | NR1D2 | 9975 | RNAseq | -0.8458 | 0.1562 | |
| SRP008496 | NR1D2 | 9975 | RNAseq | -0.6483 | 0.0617 | |
| SRP064894 | NR1D2 | 9975 | RNAseq | -1.0712 | 0.0007 | |
| SRP133303 | NR1D2 | 9975 | RNAseq | -0.2028 | 0.4891 | |
| SRP159526 | NR1D2 | 9975 | RNAseq | -0.2913 | 0.3991 | |
| SRP193095 | NR1D2 | 9975 | RNAseq | -0.9738 | 0.0000 | |
| SRP219564 | NR1D2 | 9975 | RNAseq | -0.6707 | 0.2081 | |
| TCGA | NR1D2 | 9975 | RNAseq | -0.0172 | 0.8090 |
Upregulated datasets: 0; Downregulated datasets: 2.
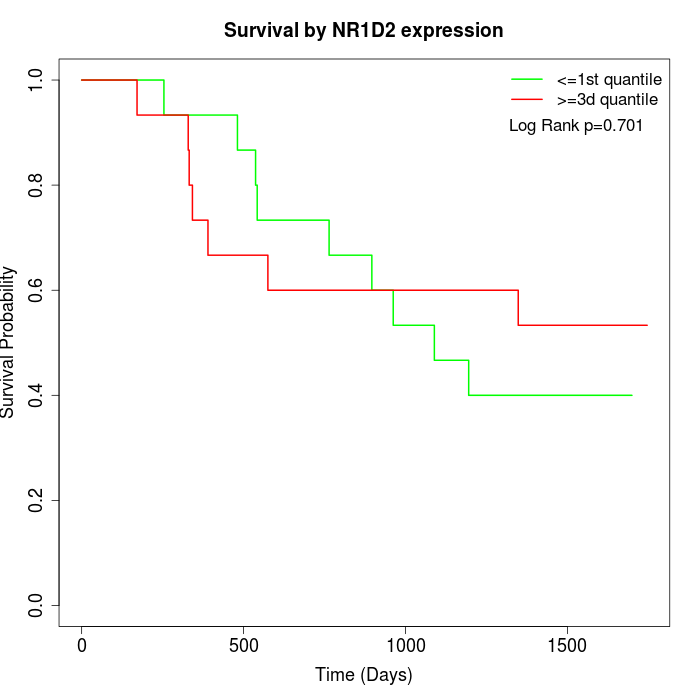
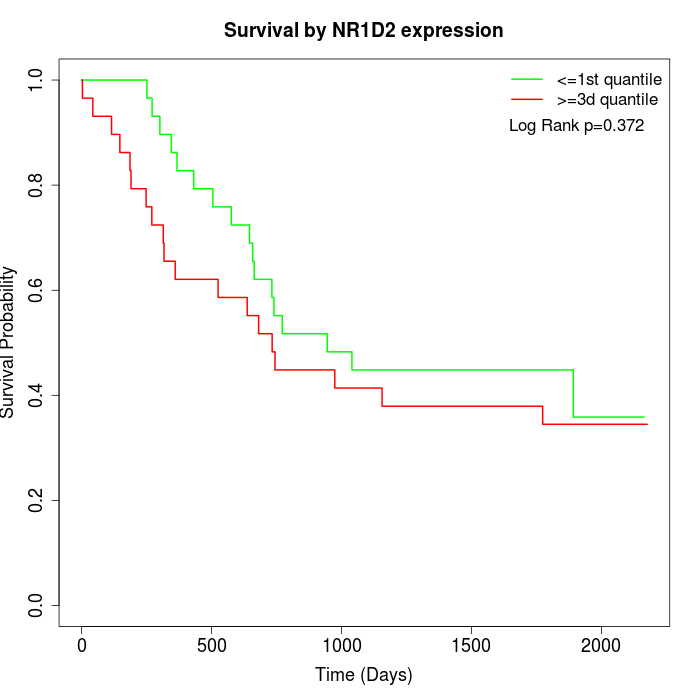
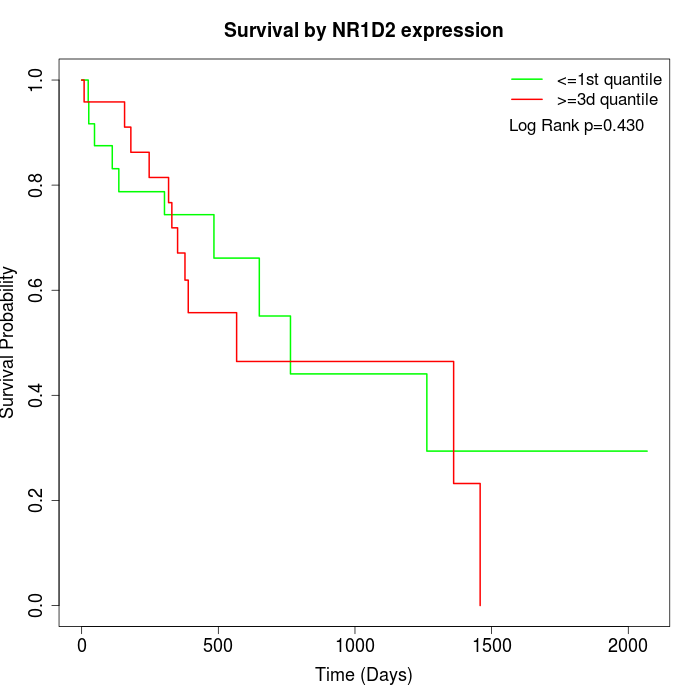
Survival by NR1D2 expression:
Note: Click image to view full size file.
Copy number change of NR1D2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NR1D2 | 9975 | 0 | 18 | 12 | |
| GSE20123 | NR1D2 | 9975 | 0 | 18 | 12 | |
| GSE43470 | NR1D2 | 9975 | 0 | 20 | 23 | |
| GSE46452 | NR1D2 | 9975 | 2 | 17 | 40 | |
| GSE47630 | NR1D2 | 9975 | 1 | 24 | 15 | |
| GSE54993 | NR1D2 | 9975 | 6 | 2 | 62 | |
| GSE54994 | NR1D2 | 9975 | 1 | 32 | 20 | |
| GSE60625 | NR1D2 | 9975 | 5 | 0 | 6 | |
| GSE74703 | NR1D2 | 9975 | 0 | 17 | 19 | |
| GSE74704 | NR1D2 | 9975 | 0 | 12 | 8 | |
| TCGA | NR1D2 | 9975 | 0 | 69 | 27 |
Total number of gains: 15; Total number of losses: 229; Total Number of normals: 244.
Somatic mutations of NR1D2:
Generating mutation plots.
Highly correlated genes for NR1D2:
Showing top 20/384 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NR1D2 | C5orf51 | 0.741929 | 3 | 0 | 3 |
| NR1D2 | HCFC2 | 0.69596 | 3 | 0 | 3 |
| NR1D2 | ARFGAP2 | 0.690059 | 3 | 0 | 3 |
| NR1D2 | SLC35B3 | 0.685021 | 3 | 0 | 3 |
| NR1D2 | VPS26B | 0.684381 | 3 | 0 | 3 |
| NR1D2 | NAA35 | 0.683408 | 3 | 0 | 3 |
| NR1D2 | OSBPL8 | 0.677931 | 3 | 0 | 3 |
| NR1D2 | SLAIN1 | 0.67753 | 3 | 0 | 3 |
| NR1D2 | ZNF155 | 0.677429 | 3 | 0 | 3 |
| NR1D2 | TAGAP | 0.671556 | 3 | 0 | 3 |
| NR1D2 | PANK1 | 0.661809 | 3 | 0 | 3 |
| NR1D2 | RAB21 | 0.660878 | 3 | 0 | 3 |
| NR1D2 | FOXP4 | 0.660206 | 3 | 0 | 3 |
| NR1D2 | CCIN | 0.658138 | 3 | 0 | 3 |
| NR1D2 | NUB1 | 0.653704 | 3 | 0 | 3 |
| NR1D2 | PCGF5 | 0.653654 | 3 | 0 | 3 |
| NR1D2 | CBLL1 | 0.65268 | 3 | 0 | 3 |
| NR1D2 | NR1H2 | 0.651715 | 3 | 0 | 3 |
| NR1D2 | PER3 | 0.650464 | 12 | 0 | 8 |
| NR1D2 | TOPORS | 0.649509 | 4 | 0 | 3 |
For details and further investigation, click here