| Full name: ornithine aminotransferase | Alias Symbol: HOGA | ||
| Type: protein-coding gene | Cytoband: 10q26.13 | ||
| Entrez ID: 4942 | HGNC ID: HGNC:8091 | Ensembl Gene: ENSG00000065154 | OMIM ID: 613349 |
| Drug and gene relationship at DGIdb | |||
Expression of OAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | OAT | 4942 | 201599_at | -0.4941 | 0.0403 | |
| GSE20347 | OAT | 4942 | 201599_at | -0.1995 | 0.3008 | |
| GSE23400 | OAT | 4942 | 201599_at | -0.1960 | 0.0400 | |
| GSE26886 | OAT | 4942 | 201599_at | -1.1131 | 0.0001 | |
| GSE29001 | OAT | 4942 | 201599_at | -0.1777 | 0.5393 | |
| GSE38129 | OAT | 4942 | 201599_at | -0.2243 | 0.0976 | |
| GSE45670 | OAT | 4942 | 201599_at | -0.5124 | 0.0009 | |
| GSE53622 | OAT | 4942 | 60983 | -0.7078 | 0.0000 | |
| GSE53624 | OAT | 4942 | 60983 | -0.5818 | 0.0000 | |
| GSE63941 | OAT | 4942 | 201599_at | -0.3342 | 0.5151 | |
| GSE77861 | OAT | 4942 | 201599_at | -1.0717 | 0.0087 | |
| GSE97050 | OAT | 4942 | A_23_P98092 | -0.1282 | 0.5127 | |
| SRP007169 | OAT | 4942 | RNAseq | -1.2265 | 0.0380 | |
| SRP008496 | OAT | 4942 | RNAseq | -1.1005 | 0.0011 | |
| SRP064894 | OAT | 4942 | RNAseq | -0.8100 | 0.0000 | |
| SRP133303 | OAT | 4942 | RNAseq | -0.4594 | 0.0001 | |
| SRP159526 | OAT | 4942 | RNAseq | -0.5641 | 0.1027 | |
| SRP193095 | OAT | 4942 | RNAseq | -1.1847 | 0.0000 | |
| SRP219564 | OAT | 4942 | RNAseq | -0.4422 | 0.1431 | |
| TCGA | OAT | 4942 | RNAseq | -0.0953 | 0.0523 |
Upregulated datasets: 0; Downregulated datasets: 5.
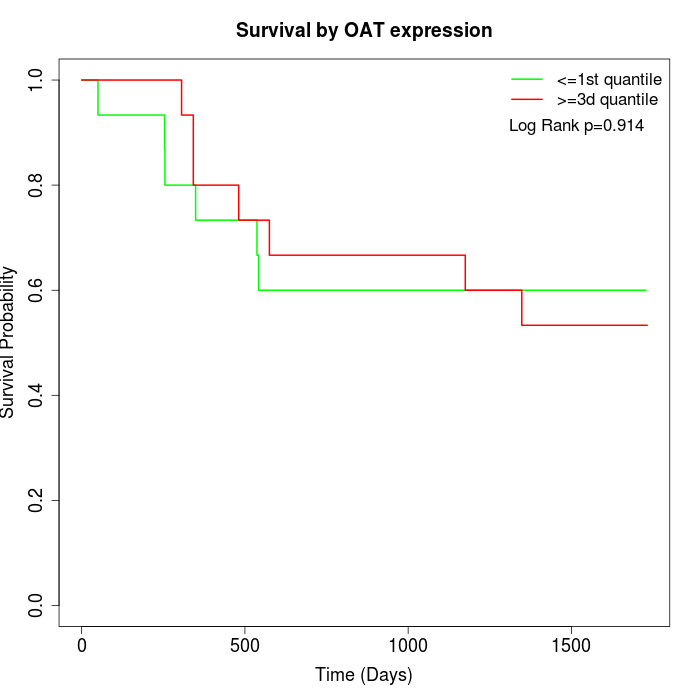
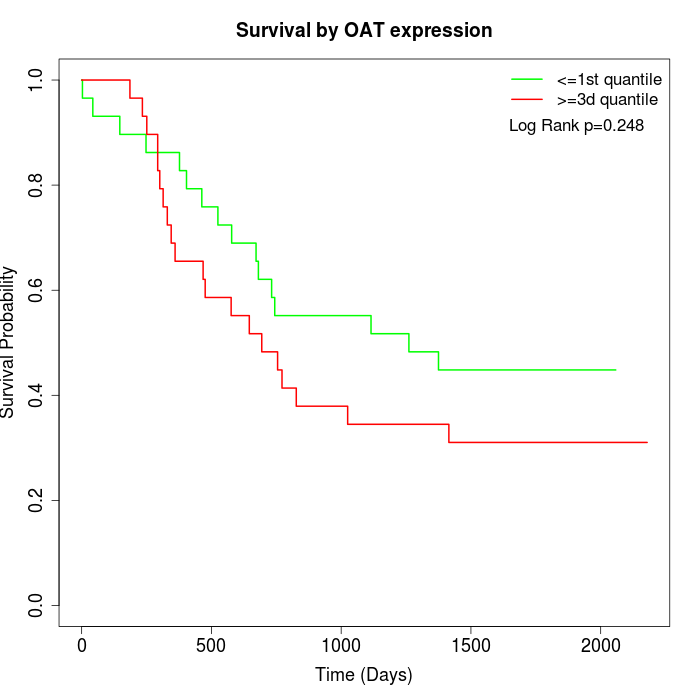
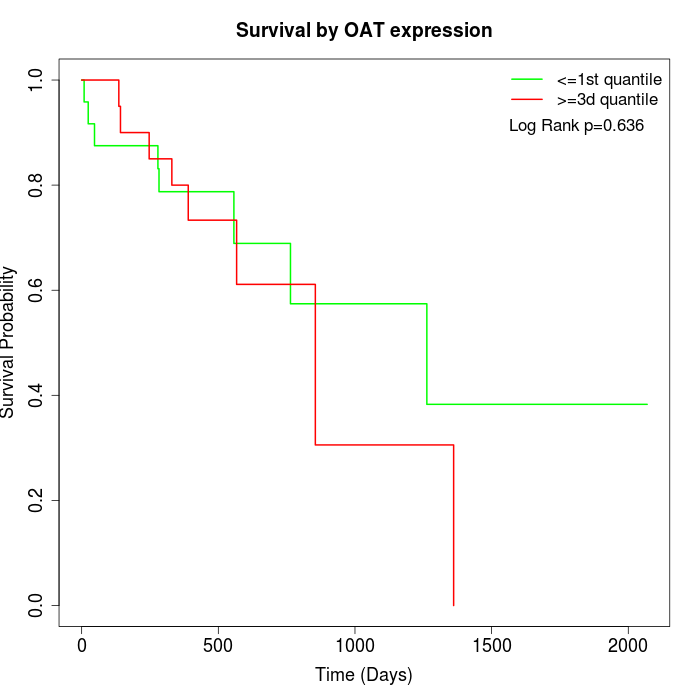
Survival by OAT expression:
Note: Click image to view full size file.
Copy number change of OAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | OAT | 4942 | 1 | 6 | 23 | |
| GSE20123 | OAT | 4942 | 1 | 6 | 23 | |
| GSE43470 | OAT | 4942 | 0 | 10 | 33 | |
| GSE46452 | OAT | 4942 | 0 | 11 | 48 | |
| GSE47630 | OAT | 4942 | 2 | 14 | 24 | |
| GSE54993 | OAT | 4942 | 8 | 1 | 61 | |
| GSE54994 | OAT | 4942 | 2 | 8 | 43 | |
| GSE60625 | OAT | 4942 | 0 | 0 | 11 | |
| GSE74703 | OAT | 4942 | 0 | 6 | 30 | |
| GSE74704 | OAT | 4942 | 1 | 2 | 17 | |
| TCGA | OAT | 4942 | 5 | 27 | 64 |
Total number of gains: 20; Total number of losses: 91; Total Number of normals: 377.
Somatic mutations of OAT:
Generating mutation plots.
Highly correlated genes for OAT:
Showing top 20/1348 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| OAT | OPHN1 | 0.80259 | 3 | 0 | 3 |
| OAT | TBC1D20 | 0.801174 | 3 | 0 | 3 |
| OAT | CYP4V2 | 0.778494 | 3 | 0 | 3 |
| OAT | EIF4G2 | 0.764896 | 3 | 0 | 3 |
| OAT | AIFM1 | 0.75344 | 3 | 0 | 3 |
| OAT | SRR | 0.745971 | 4 | 0 | 4 |
| OAT | RNF14 | 0.729759 | 4 | 0 | 4 |
| OAT | FBXW5 | 0.728291 | 4 | 0 | 4 |
| OAT | TYSND1 | 0.724726 | 3 | 0 | 3 |
| OAT | TBCA | 0.72442 | 3 | 0 | 3 |
| OAT | GNG5 | 0.724387 | 3 | 0 | 3 |
| OAT | DNAJB9 | 0.720914 | 3 | 0 | 3 |
| OAT | PDK2 | 0.713127 | 3 | 0 | 3 |
| OAT | TMEM59 | 0.710254 | 6 | 0 | 5 |
| OAT | SAMM50 | 0.710228 | 3 | 0 | 3 |
| OAT | WBP2 | 0.706978 | 3 | 0 | 3 |
| OAT | KAT2B | 0.705555 | 7 | 0 | 7 |
| OAT | THAP2 | 0.705437 | 3 | 0 | 3 |
| OAT | MBNL3 | 0.705018 | 4 | 0 | 4 |
| OAT | ZFC3H1 | 0.704337 | 3 | 0 | 3 |
For details and further investigation, click here