| Full name: poly(A) binding protein cytoplasmic 1 | Alias Symbol: PABP1|PABPL1 | ||
| Type: protein-coding gene | Cytoband: 8q22.3 | ||
| Entrez ID: 26986 | HGNC ID: HGNC:8554 | Ensembl Gene: ENSG00000070756 | OMIM ID: 604679 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PABPC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PABPC1 | 26986 | 215157_x_at | 0.1649 | 0.6173 | |
| GSE20347 | PABPC1 | 26986 | 215157_x_at | -0.2165 | 0.0531 | |
| GSE23400 | PABPC1 | 26986 | 215157_x_at | -0.2243 | 0.0019 | |
| GSE26886 | PABPC1 | 26986 | 215157_x_at | -0.5266 | 0.0014 | |
| GSE29001 | PABPC1 | 26986 | 215157_x_at | -0.2029 | 0.2095 | |
| GSE38129 | PABPC1 | 26986 | 215157_x_at | -0.0519 | 0.6639 | |
| GSE45670 | PABPC1 | 26986 | 215157_x_at | -0.0688 | 0.3820 | |
| GSE53622 | PABPC1 | 26986 | 59456 | -0.3231 | 0.0022 | |
| GSE53624 | PABPC1 | 26986 | 59456 | -0.3600 | 0.0000 | |
| GSE63941 | PABPC1 | 26986 | 215157_x_at | 0.2167 | 0.4305 | |
| GSE77861 | PABPC1 | 26986 | 215157_x_at | -0.2929 | 0.0582 | |
| GSE97050 | PABPC1 | 26986 | A_23_P82693 | -0.1734 | 0.6146 | |
| SRP007169 | PABPC1 | 26986 | RNAseq | -2.2179 | 0.0000 | |
| SRP008496 | PABPC1 | 26986 | RNAseq | -1.9841 | 0.0000 | |
| SRP064894 | PABPC1 | 26986 | RNAseq | -1.1680 | 0.0000 | |
| SRP133303 | PABPC1 | 26986 | RNAseq | -0.8661 | 0.0000 | |
| SRP159526 | PABPC1 | 26986 | RNAseq | -1.0083 | 0.0002 | |
| SRP193095 | PABPC1 | 26986 | RNAseq | -1.2484 | 0.0000 | |
| SRP219564 | PABPC1 | 26986 | RNAseq | -0.8092 | 0.2102 | |
| TCGA | PABPC1 | 26986 | RNAseq | 0.1121 | 0.0096 |
Upregulated datasets: 0; Downregulated datasets: 5.
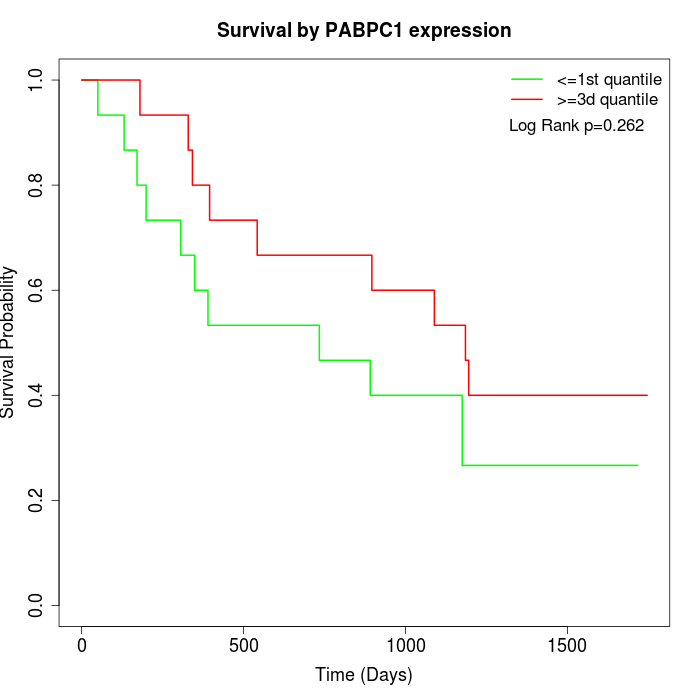
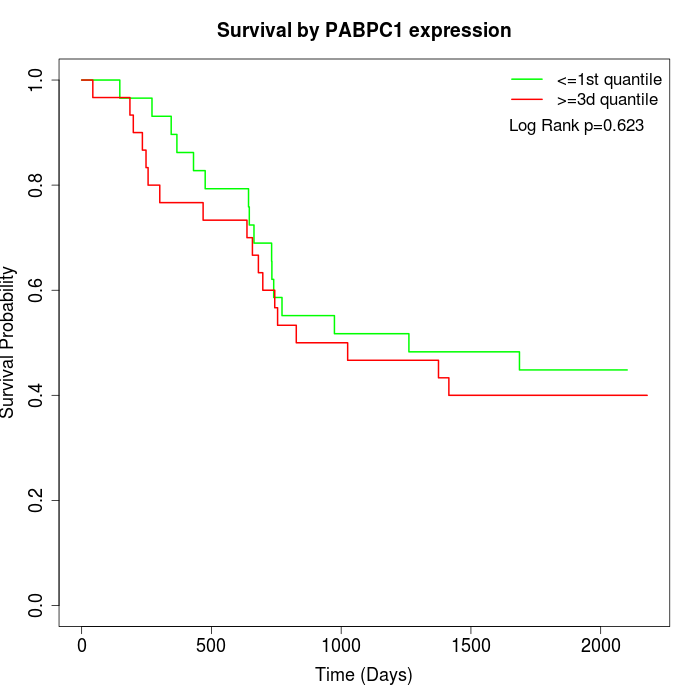
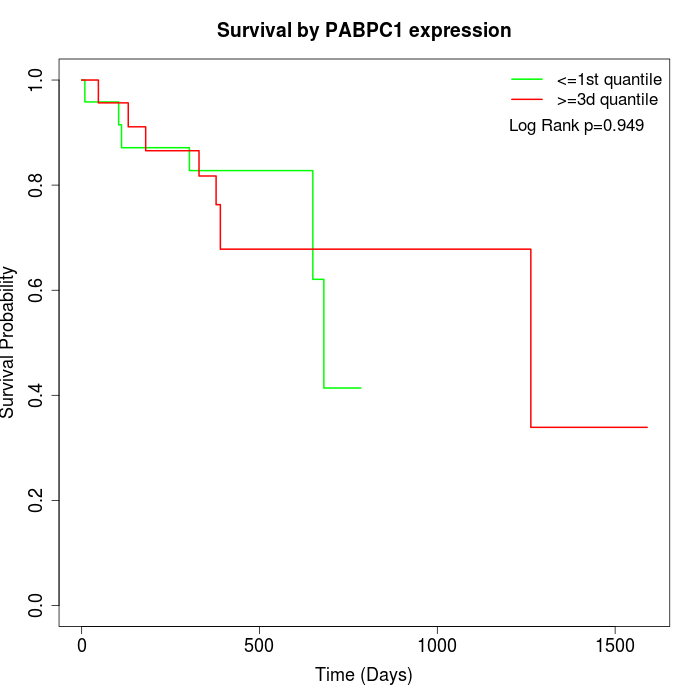
Survival by PABPC1 expression:
Note: Click image to view full size file.
Copy number change of PABPC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PABPC1 | 26986 | 17 | 0 | 13 | |
| GSE20123 | PABPC1 | 26986 | 17 | 0 | 13 | |
| GSE43470 | PABPC1 | 26986 | 22 | 0 | 21 | |
| GSE46452 | PABPC1 | 26986 | 24 | 0 | 35 | |
| GSE47630 | PABPC1 | 26986 | 24 | 0 | 16 | |
| GSE54993 | PABPC1 | 26986 | 0 | 20 | 50 | |
| GSE54994 | PABPC1 | 26986 | 38 | 1 | 14 | |
| GSE60625 | PABPC1 | 26986 | 0 | 4 | 7 | |
| GSE74703 | PABPC1 | 26986 | 19 | 0 | 17 | |
| GSE74704 | PABPC1 | 26986 | 12 | 0 | 8 | |
| TCGA | PABPC1 | 26986 | 59 | 1 | 36 |
Total number of gains: 232; Total number of losses: 26; Total Number of normals: 230.
Somatic mutations of PABPC1:
Generating mutation plots.
Highly correlated genes for PABPC1:
Showing top 20/295 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PABPC1 | PABPC3 | 0.868324 | 13 | 0 | 13 |
| PABPC1 | CPNE3 | 0.679291 | 12 | 0 | 11 |
| PABPC1 | CCDC96 | 0.657732 | 3 | 0 | 3 |
| PABPC1 | LCORL | 0.656141 | 4 | 0 | 3 |
| PABPC1 | MST1 | 0.649765 | 5 | 0 | 4 |
| PABPC1 | CSTA | 0.634058 | 4 | 0 | 4 |
| PABPC1 | ZBTB42 | 0.634004 | 3 | 0 | 3 |
| PABPC1 | SCRIB | 0.623702 | 10 | 0 | 7 |
| PABPC1 | MBD6 | 0.61983 | 3 | 0 | 3 |
| PABPC1 | BCAP31 | 0.619427 | 4 | 0 | 3 |
| PABPC1 | POR | 0.614788 | 3 | 0 | 3 |
| PABPC1 | ESRP1 | 0.613589 | 12 | 0 | 9 |
| PABPC1 | ZNF813 | 0.613227 | 3 | 0 | 3 |
| PABPC1 | CNFN | 0.612972 | 5 | 0 | 4 |
| PABPC1 | EPHA10 | 0.611056 | 3 | 0 | 3 |
| PABPC1 | PRSS27 | 0.606289 | 6 | 0 | 5 |
| PABPC1 | AIF1L | 0.605172 | 5 | 0 | 5 |
| PABPC1 | RNF39 | 0.603258 | 6 | 0 | 4 |
| PABPC1 | KRT13 | 0.601231 | 6 | 0 | 4 |
| PABPC1 | ACSM3 | 0.599799 | 4 | 0 | 3 |
For details and further investigation, click here