| Full name: scribble planar cell polarity protein | Alias Symbol: KIAA0147|SCRB1|Vartul | ||
| Type: protein-coding gene | Cytoband: 8q24.3 | ||
| Entrez ID: 23513 | HGNC ID: HGNC:30377 | Ensembl Gene: ENSG00000180900 | OMIM ID: 607733 |
| Drug and gene relationship at DGIdb | |||
SCRIB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway |
Expression of SCRIB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SCRIB | 23513 | 212556_at | 0.3168 | 0.6692 | |
| GSE20347 | SCRIB | 23513 | 212556_at | -0.3313 | 0.0562 | |
| GSE23400 | SCRIB | 23513 | 212556_at | -0.2883 | 0.0041 | |
| GSE26886 | SCRIB | 23513 | 212556_at | 0.1828 | 0.2769 | |
| GSE29001 | SCRIB | 23513 | 212556_at | -0.6240 | 0.0241 | |
| GSE38129 | SCRIB | 23513 | 212556_at | -0.0639 | 0.7463 | |
| GSE45670 | SCRIB | 23513 | 212556_at | 0.1178 | 0.4555 | |
| GSE53622 | SCRIB | 23513 | 115748 | -0.4859 | 0.0000 | |
| GSE53624 | SCRIB | 23513 | 115748 | -0.4730 | 0.0000 | |
| GSE63941 | SCRIB | 23513 | 212556_at | 0.8191 | 0.2178 | |
| GSE77861 | SCRIB | 23513 | 212556_at | -0.1736 | 0.3747 | |
| GSE97050 | SCRIB | 23513 | A_23_P159039 | -0.1499 | 0.4428 | |
| SRP007169 | SCRIB | 23513 | RNAseq | -0.6629 | 0.0855 | |
| SRP008496 | SCRIB | 23513 | RNAseq | -0.7839 | 0.0024 | |
| SRP064894 | SCRIB | 23513 | RNAseq | -0.3548 | 0.1924 | |
| SRP133303 | SCRIB | 23513 | RNAseq | -0.6769 | 0.0005 | |
| SRP159526 | SCRIB | 23513 | RNAseq | -0.3584 | 0.2565 | |
| SRP193095 | SCRIB | 23513 | RNAseq | -0.3368 | 0.0499 | |
| SRP219564 | SCRIB | 23513 | RNAseq | -0.0264 | 0.9662 | |
| TCGA | SCRIB | 23513 | RNAseq | 0.1662 | 0.0009 |
Upregulated datasets: 0; Downregulated datasets: 0.
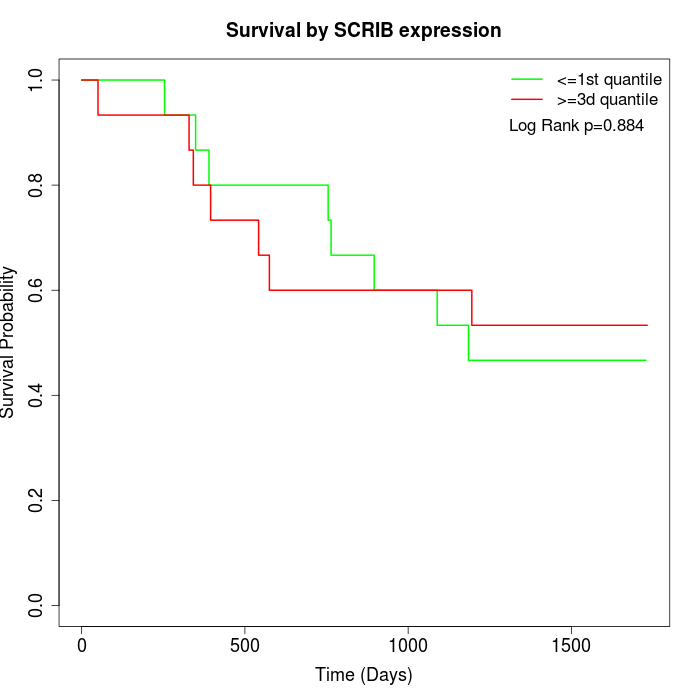
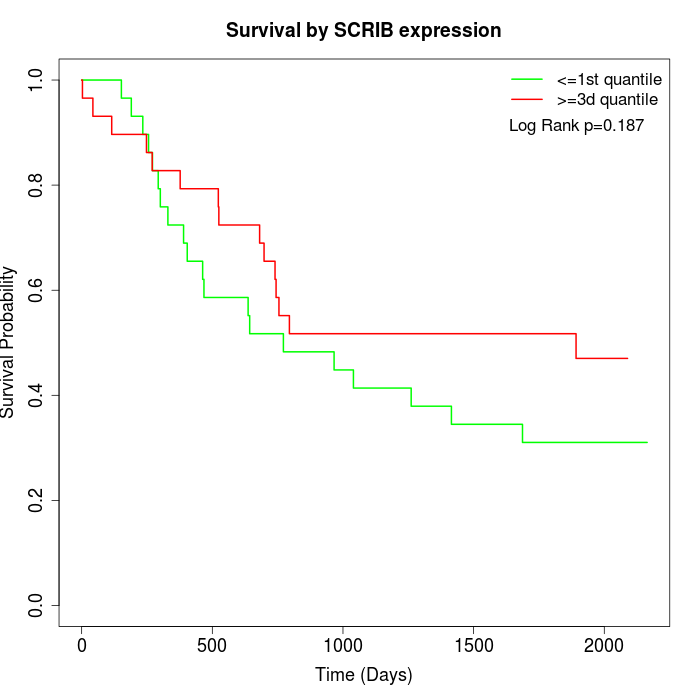
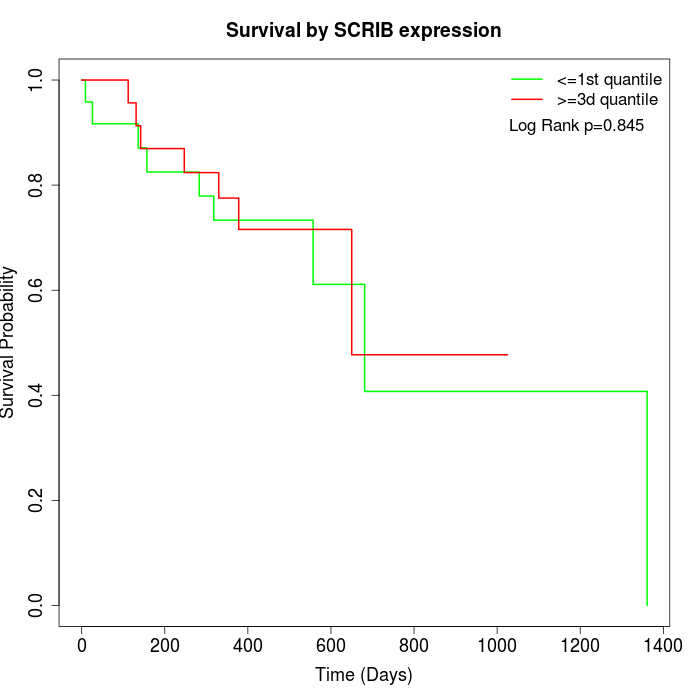
Survival by SCRIB expression:
Note: Click image to view full size file.
Copy number change of SCRIB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SCRIB | 23513 | 17 | 1 | 12 | |
| GSE20123 | SCRIB | 23513 | 17 | 1 | 12 | |
| GSE43470 | SCRIB | 23513 | 16 | 2 | 25 | |
| GSE46452 | SCRIB | 23513 | 27 | 0 | 32 | |
| GSE47630 | SCRIB | 23513 | 23 | 1 | 16 | |
| GSE54993 | SCRIB | 23513 | 0 | 25 | 45 | |
| GSE54994 | SCRIB | 23513 | 39 | 1 | 13 | |
| GSE60625 | SCRIB | 23513 | 3 | 4 | 4 | |
| GSE74703 | SCRIB | 23513 | 14 | 1 | 21 | |
| GSE74704 | SCRIB | 23513 | 10 | 0 | 10 | |
| TCGA | SCRIB | 23513 | 64 | 2 | 30 |
Total number of gains: 230; Total number of losses: 38; Total Number of normals: 220.
Somatic mutations of SCRIB:
Generating mutation plots.
Highly correlated genes for SCRIB:
Showing top 20/375 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SCRIB | NCCRP1 | 0.675998 | 3 | 0 | 3 |
| SCRIB | PABPC3 | 0.670785 | 10 | 0 | 9 |
| SCRIB | CPNE3 | 0.664938 | 8 | 0 | 8 |
| SCRIB | SH3D21 | 0.664086 | 3 | 0 | 3 |
| SCRIB | DENND1B | 0.660948 | 4 | 0 | 4 |
| SCRIB | ESRP1 | 0.660079 | 8 | 0 | 6 |
| SCRIB | SPRR2F | 0.627663 | 3 | 0 | 3 |
| SCRIB | MROH6 | 0.626425 | 5 | 0 | 4 |
| SCRIB | PABPC1 | 0.623702 | 10 | 0 | 7 |
| SCRIB | LLGL2 | 0.621217 | 9 | 0 | 9 |
| SCRIB | CNFN | 0.617353 | 3 | 0 | 3 |
| SCRIB | MAPK13 | 0.616385 | 8 | 0 | 6 |
| SCRIB | CSTB | 0.61334 | 6 | 0 | 6 |
| SCRIB | RAD9A | 0.60884 | 4 | 0 | 3 |
| SCRIB | DGKA | 0.6069 | 9 | 0 | 6 |
| SCRIB | KIFC2 | 0.606188 | 5 | 0 | 4 |
| SCRIB | DGAT1 | 0.604742 | 11 | 0 | 7 |
| SCRIB | STAP2 | 0.604682 | 5 | 0 | 4 |
| SCRIB | KAZN | 0.599304 | 10 | 0 | 9 |
| SCRIB | RAPGEFL1 | 0.598511 | 9 | 0 | 8 |
For details and further investigation, click here