| Full name: PAS domain containing repressor 1 | Alias Symbol: CT63 | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 139135 | HGNC ID: HGNC:20686 | Ensembl Gene: ENSG00000166049 | OMIM ID: 300993 |
| Drug and gene relationship at DGIdb | |||
Expression of PASD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PASD1 | 139135 | 240687_at | 0.3614 | 0.3841 | |
| GSE26886 | PASD1 | 139135 | 240687_at | 0.3012 | 0.0003 | |
| GSE45670 | PASD1 | 139135 | 240687_at | 0.1529 | 0.3010 | |
| GSE53622 | PASD1 | 139135 | 98710 | 0.2544 | 0.0547 | |
| GSE53624 | PASD1 | 139135 | 98710 | 0.2453 | 0.1547 | |
| GSE63941 | PASD1 | 139135 | 240687_at | 0.3007 | 0.6824 | |
| GSE77861 | PASD1 | 139135 | 240687_at | -0.0567 | 0.6839 | |
| TCGA | PASD1 | 139135 | RNAseq | 2.4912 | 0.2451 |
Upregulated datasets: 0; Downregulated datasets: 0.
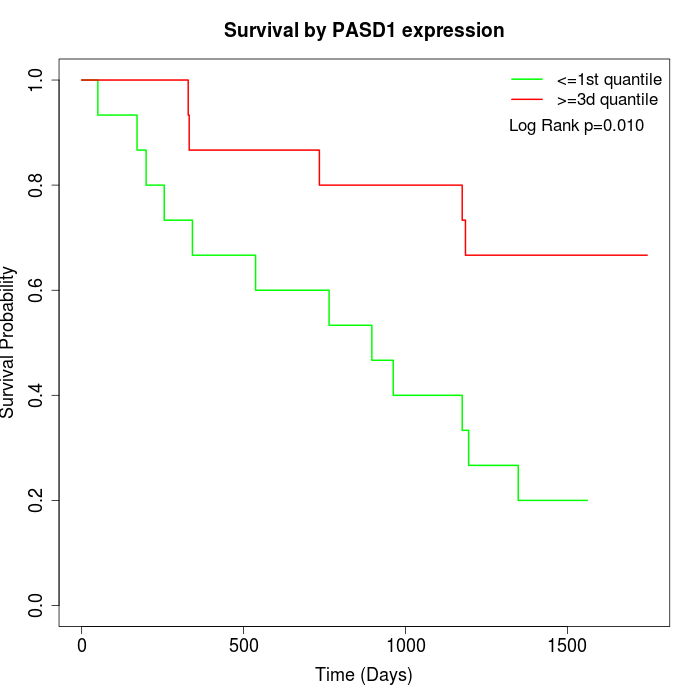
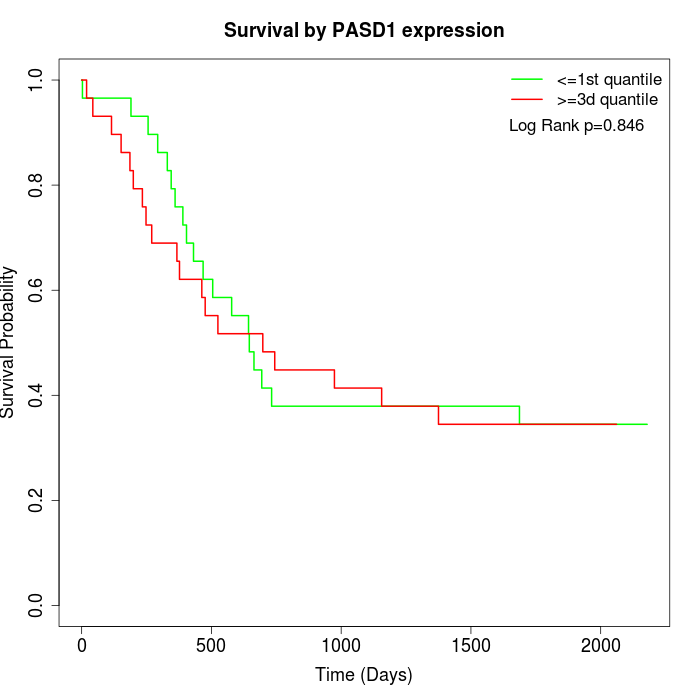
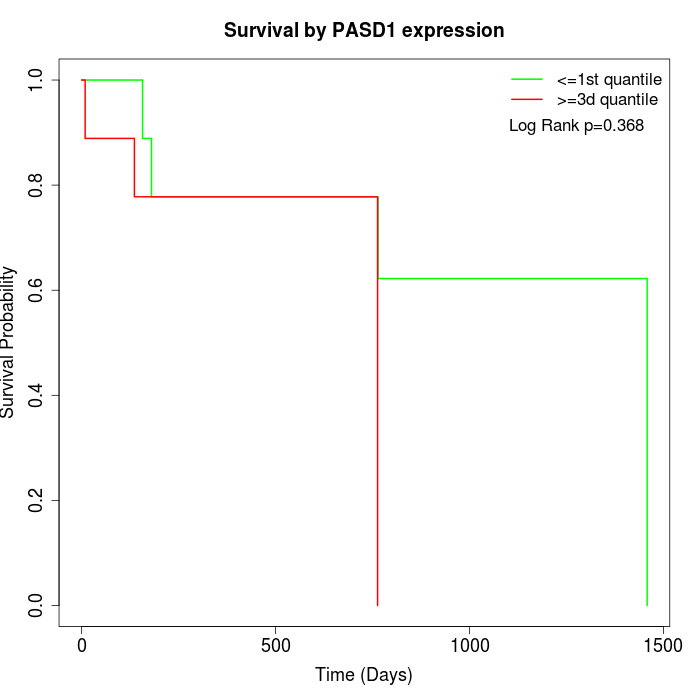
Survival by PASD1 expression:
Note: Click image to view full size file.
Copy number change of PASD1:
No record found for this gene.
Somatic mutations of PASD1:
Generating mutation plots.
Highly correlated genes for PASD1:
Showing top 20/31 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PASD1 | CTCFL | 0.799324 | 3 | 0 | 3 |
| PASD1 | LIM2 | 0.734718 | 3 | 0 | 3 |
| PASD1 | FAM230C | 0.734288 | 3 | 0 | 3 |
| PASD1 | CMTM1 | 0.717007 | 3 | 0 | 3 |
| PASD1 | KLK14 | 0.715187 | 3 | 0 | 3 |
| PASD1 | PRIM2 | 0.69456 | 3 | 0 | 3 |
| PASD1 | ACTR5 | 0.693806 | 3 | 0 | 3 |
| PASD1 | GPR150 | 0.686087 | 3 | 0 | 3 |
| PASD1 | MCEMP1 | 0.669661 | 3 | 0 | 3 |
| PASD1 | CCDC28B | 0.663941 | 3 | 0 | 3 |
| PASD1 | PSKH1 | 0.634674 | 3 | 0 | 3 |
| PASD1 | RCE1 | 0.624865 | 3 | 0 | 3 |
| PASD1 | CPLX3 | 0.614996 | 3 | 0 | 3 |
| PASD1 | KPTN | 0.612438 | 3 | 0 | 3 |
| PASD1 | PAGE1 | 0.608488 | 4 | 0 | 4 |
| PASD1 | GLTPD2 | 0.604334 | 3 | 0 | 3 |
| PASD1 | HOXB6 | 0.603688 | 3 | 0 | 3 |
| PASD1 | ICAM5 | 0.602951 | 3 | 0 | 3 |
| PASD1 | ELFN1 | 0.600938 | 3 | 0 | 3 |
| PASD1 | SRA1 | 0.599414 | 3 | 0 | 3 |
For details and further investigation, click here