| Full name: prostate cancer associated transcript 18 | Alias Symbol: LINC01092 | ||
| Type: non-coding RNA | Cytoband: 18q11.2 | ||
| Entrez ID: 728606 | HGNC ID: HGNC:49211 | Ensembl Gene: ENSG00000265369 | OMIM ID: 617647 |
| Drug and gene relationship at DGIdb | |||
Expression of PCAT18:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PCAT18 | 728606 | 1559276_at | -0.1655 | 0.4284 | |
| GSE26886 | PCAT18 | 728606 | 1559276_at | 0.0020 | 0.9834 | |
| GSE45670 | PCAT18 | 728606 | 1559276_at | -0.0244 | 0.8170 | |
| GSE53622 | PCAT18 | 728606 | 3034 | -0.5979 | 0.0005 | |
| GSE53624 | PCAT18 | 728606 | 3034 | -0.8846 | 0.0000 | |
| GSE63941 | PCAT18 | 728606 | 1559276_at | 0.0353 | 0.8263 | |
| GSE77861 | PCAT18 | 728606 | 1559276_at | -0.1442 | 0.0555 | |
| SRP064894 | PCAT18 | 728606 | RNAseq | -2.8056 | 0.0000 | |
| SRP133303 | PCAT18 | 728606 | RNAseq | -0.6387 | 0.0006 | |
| SRP159526 | PCAT18 | 728606 | RNAseq | -2.0859 | 0.0032 | |
| SRP193095 | PCAT18 | 728606 | RNAseq | -0.6572 | 0.0240 |
Upregulated datasets: 0; Downregulated datasets: 2.
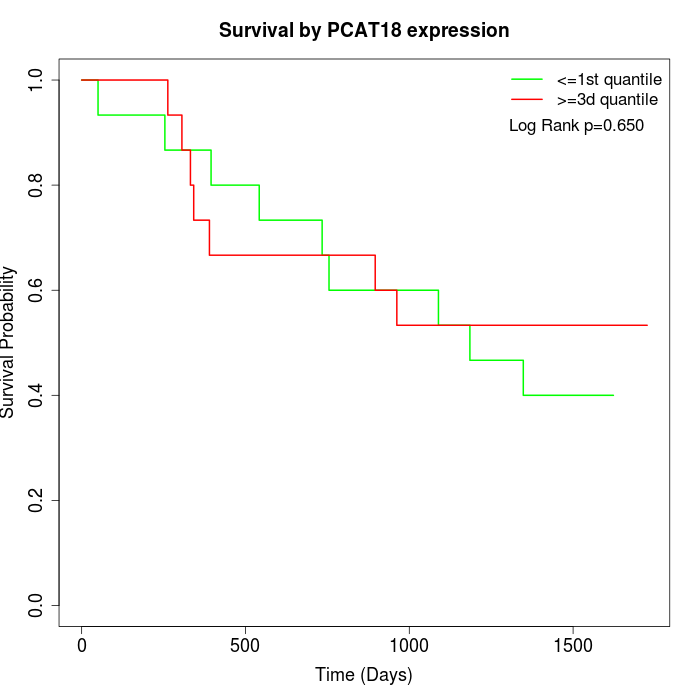
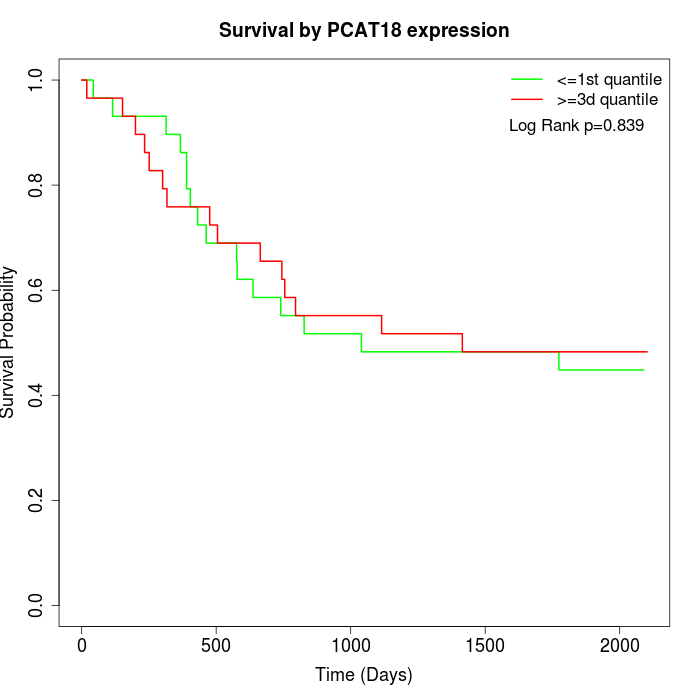
Survival by PCAT18 expression:
Note: Click image to view full size file.
Copy number change of PCAT18:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PCAT18 | 728606 | 2 | 5 | 23 | |
| GSE20123 | PCAT18 | 728606 | 2 | 5 | 23 | |
| GSE43470 | PCAT18 | 728606 | 0 | 2 | 41 | |
| GSE46452 | PCAT18 | 728606 | 1 | 25 | 33 | |
| GSE47630 | PCAT18 | 728606 | 5 | 19 | 16 | |
| GSE54993 | PCAT18 | 728606 | 7 | 1 | 62 | |
| GSE54994 | PCAT18 | 728606 | 3 | 14 | 36 | |
| GSE60625 | PCAT18 | 728606 | 0 | 4 | 7 | |
| GSE74703 | PCAT18 | 728606 | 0 | 2 | 34 | |
| GSE74704 | PCAT18 | 728606 | 1 | 4 | 15 |
Total number of gains: 21; Total number of losses: 81; Total Number of normals: 290.
Somatic mutations of PCAT18:
Generating mutation plots.
Highly correlated genes for PCAT18:
Showing top 20/36 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PCAT18 | TSPAN8 | 0.669255 | 3 | 0 | 3 |
| PCAT18 | S100G | 0.66134 | 3 | 0 | 3 |
| PCAT18 | PDE10A | 0.66019 | 3 | 0 | 3 |
| PCAT18 | LGALS13 | 0.655527 | 3 | 0 | 3 |
| PCAT18 | PCA3 | 0.651034 | 3 | 0 | 3 |
| PCAT18 | P2RX1 | 0.638687 | 3 | 0 | 3 |
| PCAT18 | PROSER2-AS1 | 0.604955 | 3 | 0 | 3 |
| PCAT18 | MS4A8 | 0.604452 | 4 | 0 | 3 |
| PCAT18 | AZGP1 | 0.600348 | 3 | 0 | 3 |
| PCAT18 | ADCYAP1R1 | 0.599534 | 3 | 0 | 3 |
| PCAT18 | SLC44A4 | 0.597734 | 4 | 0 | 3 |
| PCAT18 | PROM1 | 0.597045 | 3 | 0 | 3 |
| PCAT18 | CCL23 | 0.592542 | 3 | 0 | 3 |
| PCAT18 | PRSS33 | 0.590972 | 3 | 0 | 3 |
| PCAT18 | KCNA5 | 0.587343 | 4 | 0 | 3 |
| PCAT18 | ACSM3 | 0.577674 | 4 | 0 | 3 |
| PCAT18 | TOX3 | 0.577362 | 4 | 0 | 4 |
| PCAT18 | TESC | 0.573197 | 3 | 0 | 3 |
| PCAT18 | STH | 0.564441 | 3 | 0 | 3 |
| PCAT18 | LRRC31 | 0.563695 | 4 | 0 | 3 |
For details and further investigation, click here