| Full name: C-C motif chemokine ligand 23 | Alias Symbol: Ckb-8|MPIF-1|MIP-3|CKb8 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 6368 | HGNC ID: HGNC:10622 | Ensembl Gene: ENSG00000274736 | OMIM ID: 602494 |
| Drug and gene relationship at DGIdb | |||
CCL23 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL23:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL23 | 6368 | 210549_s_at | -0.6998 | 0.1779 | |
| GSE20347 | CCL23 | 6368 | 210548_at | 0.0373 | 0.5686 | |
| GSE23400 | CCL23 | 6368 | 210548_at | 0.0003 | 0.9930 | |
| GSE26886 | CCL23 | 6368 | 210548_at | 0.1446 | 0.1469 | |
| GSE29001 | CCL23 | 6368 | 210548_at | 0.0142 | 0.9404 | |
| GSE38129 | CCL23 | 6368 | 210549_s_at | -0.3311 | 0.0273 | |
| GSE45670 | CCL23 | 6368 | 210549_s_at | -0.7445 | 0.0000 | |
| GSE63941 | CCL23 | 6368 | 210549_s_at | 0.0747 | 0.6681 | |
| GSE77861 | CCL23 | 6368 | 210549_s_at | -0.1846 | 0.0572 | |
| TCGA | CCL23 | 6368 | RNAseq | -2.2852 | 0.0035 |
Upregulated datasets: 0; Downregulated datasets: 1.
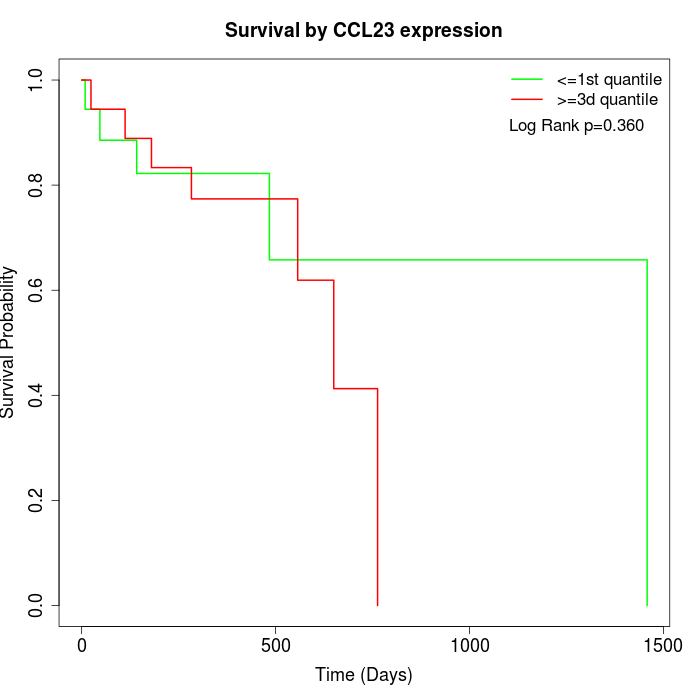
Survival by CCL23 expression:
Note: Click image to view full size file.
Copy number change of CCL23:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL23 | 6368 | 5 | 1 | 24 | |
| GSE20123 | CCL23 | 6368 | 5 | 1 | 24 | |
| GSE43470 | CCL23 | 6368 | 1 | 2 | 40 | |
| GSE46452 | CCL23 | 6368 | 34 | 0 | 25 | |
| GSE47630 | CCL23 | 6368 | 7 | 1 | 32 | |
| GSE54993 | CCL23 | 6368 | 3 | 3 | 64 | |
| GSE54994 | CCL23 | 6368 | 8 | 6 | 39 | |
| GSE60625 | CCL23 | 6368 | 4 | 0 | 7 | |
| GSE74703 | CCL23 | 6368 | 1 | 1 | 34 | |
| GSE74704 | CCL23 | 6368 | 3 | 1 | 16 | |
| TCGA | CCL23 | 6368 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of CCL23:
Generating mutation plots.
Highly correlated genes for CCL23:
Showing top 20/372 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL23 | MYRIP | 0.783217 | 3 | 0 | 3 |
| CCL23 | LYVE1 | 0.761155 | 4 | 0 | 4 |
| CCL23 | HPGDS | 0.747557 | 3 | 0 | 3 |
| CCL23 | ASPA | 0.730348 | 3 | 0 | 3 |
| CCL23 | RNASE4 | 0.726816 | 3 | 0 | 3 |
| CCL23 | RSPO1 | 0.721907 | 3 | 0 | 3 |
| CCL23 | ABCA8 | 0.718433 | 3 | 0 | 3 |
| CCL23 | ITGA8 | 0.717291 | 3 | 0 | 3 |
| CCL23 | MEOX2 | 0.710452 | 4 | 0 | 4 |
| CCL23 | PLAC9 | 0.7081 | 3 | 0 | 3 |
| CCL23 | RERGL | 0.706762 | 4 | 0 | 4 |
| CCL23 | MFAP4 | 0.704643 | 4 | 0 | 4 |
| CCL23 | LRRC2 | 0.701123 | 4 | 0 | 4 |
| CCL23 | APOD | 0.70074 | 3 | 0 | 3 |
| CCL23 | C7 | 0.697286 | 3 | 0 | 3 |
| CCL23 | FGF7 | 0.696015 | 3 | 0 | 3 |
| CCL23 | KLHDC8A | 0.693908 | 3 | 0 | 3 |
| CCL23 | F10 | 0.693308 | 4 | 0 | 4 |
| CCL23 | RCAN2 | 0.689543 | 3 | 0 | 3 |
| CCL23 | P2RY14 | 0.678593 | 3 | 0 | 3 |
For details and further investigation, click here