| Full name: phosphodiesterase 1C | Alias Symbol: Hcam3 | ||
| Type: protein-coding gene | Cytoband: 7p14.3 | ||
| Entrez ID: 5137 | HGNC ID: HGNC:8776 | Ensembl Gene: ENSG00000154678 | OMIM ID: 602987 |
| Drug and gene relationship at DGIdb | |||
PDE1C involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04740 | Olfactory transduction | |
| hsa04742 | Taste transduction | |
| hsa04924 | Renin secretion |
Expression of PDE1C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE1C | 5137 | 236344_at | -1.3844 | 0.0559 | |
| GSE20347 | PDE1C | 5137 | 216869_at | 0.1159 | 0.1638 | |
| GSE23400 | PDE1C | 5137 | 207303_at | -0.2025 | 0.0000 | |
| GSE26886 | PDE1C | 5137 | 216869_at | 0.1963 | 0.0807 | |
| GSE29001 | PDE1C | 5137 | 216869_at | -0.0307 | 0.8473 | |
| GSE38129 | PDE1C | 5137 | 216869_at | -0.2287 | 0.0445 | |
| GSE45670 | PDE1C | 5137 | 216869_at | -0.4375 | 0.0001 | |
| GSE53622 | PDE1C | 5137 | 68352 | -1.9953 | 0.0000 | |
| GSE53624 | PDE1C | 5137 | 68352 | -1.7529 | 0.0000 | |
| GSE63941 | PDE1C | 5137 | 236344_at | -2.1920 | 0.0047 | |
| GSE77861 | PDE1C | 5137 | 236344_at | -0.0680 | 0.4424 | |
| GSE97050 | PDE1C | 5137 | A_24_P206328 | -1.1602 | 0.1399 | |
| SRP064894 | PDE1C | 5137 | RNAseq | -1.5921 | 0.0000 | |
| SRP133303 | PDE1C | 5137 | RNAseq | -0.8000 | 0.0401 | |
| SRP159526 | PDE1C | 5137 | RNAseq | -2.0110 | 0.0000 | |
| SRP193095 | PDE1C | 5137 | RNAseq | 0.1312 | 0.6888 | |
| SRP219564 | PDE1C | 5137 | RNAseq | -1.3314 | 0.0535 | |
| TCGA | PDE1C | 5137 | RNAseq | -1.7237 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 6.
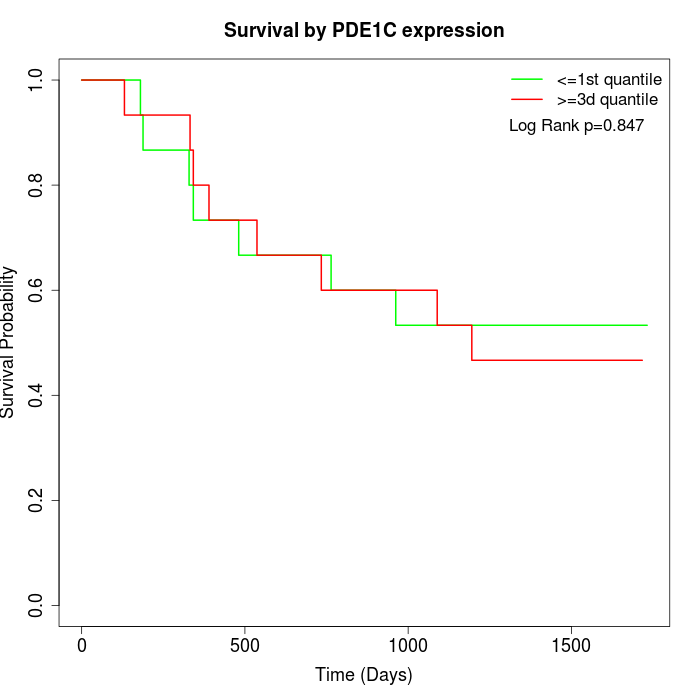
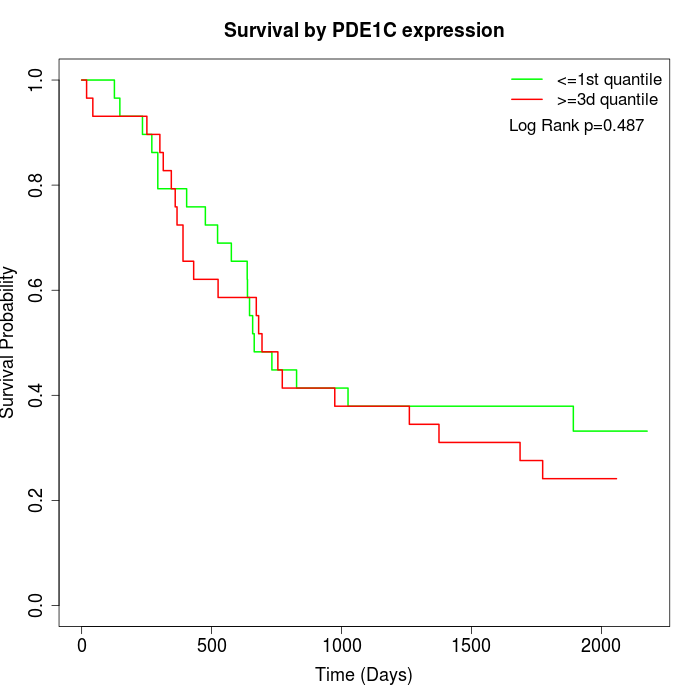
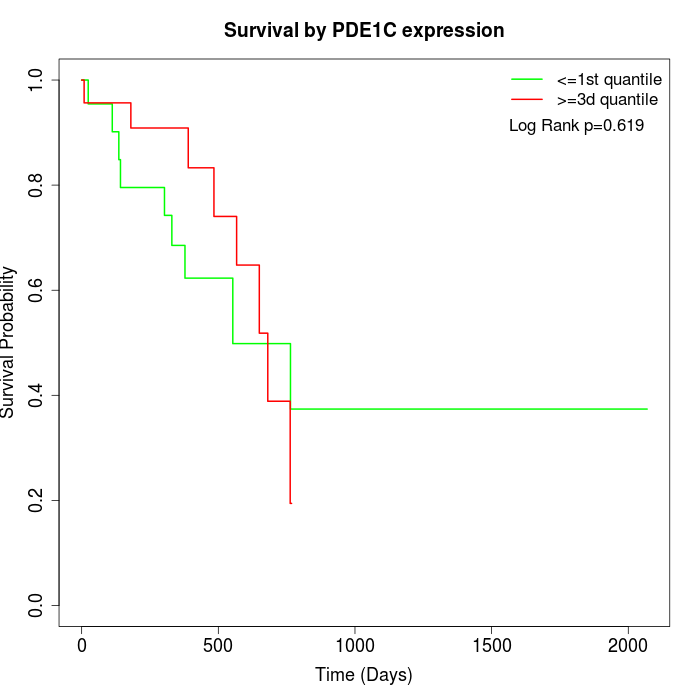
Survival by PDE1C expression:
Note: Click image to view full size file.
Copy number change of PDE1C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE1C | 5137 | 15 | 0 | 15 | |
| GSE20123 | PDE1C | 5137 | 15 | 0 | 15 | |
| GSE43470 | PDE1C | 5137 | 4 | 1 | 38 | |
| GSE46452 | PDE1C | 5137 | 11 | 1 | 47 | |
| GSE47630 | PDE1C | 5137 | 8 | 1 | 31 | |
| GSE54993 | PDE1C | 5137 | 0 | 6 | 64 | |
| GSE54994 | PDE1C | 5137 | 19 | 3 | 31 | |
| GSE60625 | PDE1C | 5137 | 0 | 0 | 11 | |
| GSE74703 | PDE1C | 5137 | 4 | 1 | 31 | |
| GSE74704 | PDE1C | 5137 | 9 | 0 | 11 | |
| TCGA | PDE1C | 5137 | 53 | 9 | 34 |
Total number of gains: 138; Total number of losses: 22; Total Number of normals: 328.
Somatic mutations of PDE1C:
Generating mutation plots.
Highly correlated genes for PDE1C:
Showing top 20/1068 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE1C | NEGR1 | 0.836084 | 6 | 0 | 6 |
| PDE1C | PLAC9 | 0.832612 | 4 | 0 | 4 |
| PDE1C | CYP21A2 | 0.829248 | 3 | 0 | 3 |
| PDE1C | CLEC3B | 0.823055 | 3 | 0 | 3 |
| PDE1C | XKR4 | 0.822211 | 5 | 0 | 5 |
| PDE1C | GABARAPL1 | 0.796257 | 3 | 0 | 3 |
| PDE1C | RASSF3 | 0.793702 | 5 | 0 | 5 |
| PDE1C | RGS7BP | 0.79315 | 5 | 0 | 5 |
| PDE1C | RBFOX3 | 0.791686 | 3 | 0 | 3 |
| PDE1C | NEXN | 0.787463 | 6 | 0 | 6 |
| PDE1C | ASB5 | 0.781608 | 6 | 0 | 6 |
| PDE1C | SORCS1 | 0.772542 | 5 | 0 | 4 |
| PDE1C | VMAC | 0.77245 | 3 | 0 | 3 |
| PDE1C | SYNPO2 | 0.768071 | 6 | 0 | 6 |
| PDE1C | HPSE2 | 0.767272 | 8 | 0 | 7 |
| PDE1C | RAB9B | 0.76667 | 6 | 0 | 5 |
| PDE1C | PRUNE2 | 0.765299 | 7 | 0 | 7 |
| PDE1C | MYOCD | 0.765193 | 7 | 0 | 6 |
| PDE1C | ADCY5 | 0.759866 | 6 | 0 | 6 |
| PDE1C | CBX7 | 0.754868 | 6 | 0 | 6 |
For details and further investigation, click here