| Full name: phosphodiesterase 4D interacting protein | Alias Symbol: KIAA0477|KIAA0454|MMGL | ||
| Type: protein-coding gene | Cytoband: 1q21.2 | ||
| Entrez ID: 9659 | HGNC ID: HGNC:15580 | Ensembl Gene: ENSG00000178104 | OMIM ID: 608117 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PDE4DIP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE4DIP | 9659 | 205872_x_at | -0.4260 | 0.4796 | |
| GSE20347 | PDE4DIP | 9659 | 205872_x_at | 0.1516 | 0.2619 | |
| GSE23400 | PDE4DIP | 9659 | 205872_x_at | -0.0754 | 0.0619 | |
| GSE26886 | PDE4DIP | 9659 | 205872_x_at | 0.5131 | 0.0093 | |
| GSE29001 | PDE4DIP | 9659 | 205872_x_at | 0.1926 | 0.4382 | |
| GSE38129 | PDE4DIP | 9659 | 205872_x_at | -0.0921 | 0.6247 | |
| GSE45670 | PDE4DIP | 9659 | 205872_x_at | -0.3217 | 0.2198 | |
| GSE53622 | PDE4DIP | 9659 | 54460 | 0.2523 | 0.1086 | |
| GSE53624 | PDE4DIP | 9659 | 54460 | 0.5685 | 0.0000 | |
| GSE63941 | PDE4DIP | 9659 | 205872_x_at | -2.6819 | 0.0003 | |
| GSE77861 | PDE4DIP | 9659 | 205872_x_at | -0.0582 | 0.6335 | |
| GSE97050 | PDE4DIP | 9659 | A_33_P3246985 | -1.2364 | 0.1782 | |
| SRP007169 | PDE4DIP | 9659 | RNAseq | 0.9684 | 0.0499 | |
| SRP008496 | PDE4DIP | 9659 | RNAseq | 1.3528 | 0.0002 | |
| SRP064894 | PDE4DIP | 9659 | RNAseq | 1.0252 | 0.0000 | |
| SRP133303 | PDE4DIP | 9659 | RNAseq | 0.6573 | 0.0024 | |
| SRP159526 | PDE4DIP | 9659 | RNAseq | 0.3735 | 0.1261 | |
| SRP193095 | PDE4DIP | 9659 | RNAseq | 0.5791 | 0.0069 | |
| SRP219564 | PDE4DIP | 9659 | RNAseq | 0.9401 | 0.1076 | |
| TCGA | PDE4DIP | 9659 | RNAseq | -0.2974 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 1.
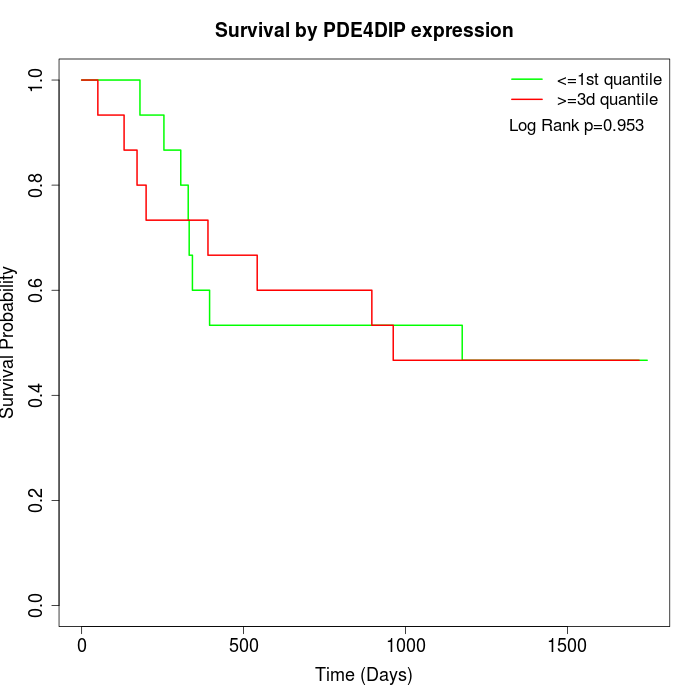
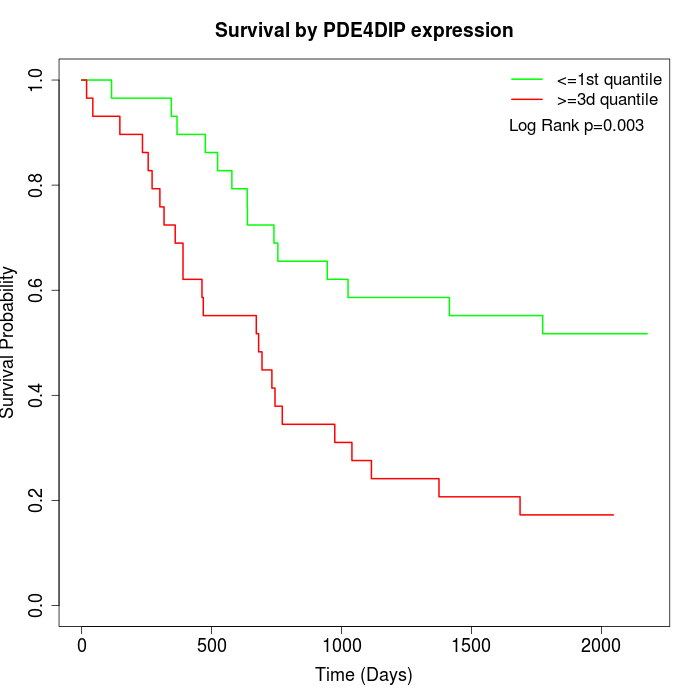
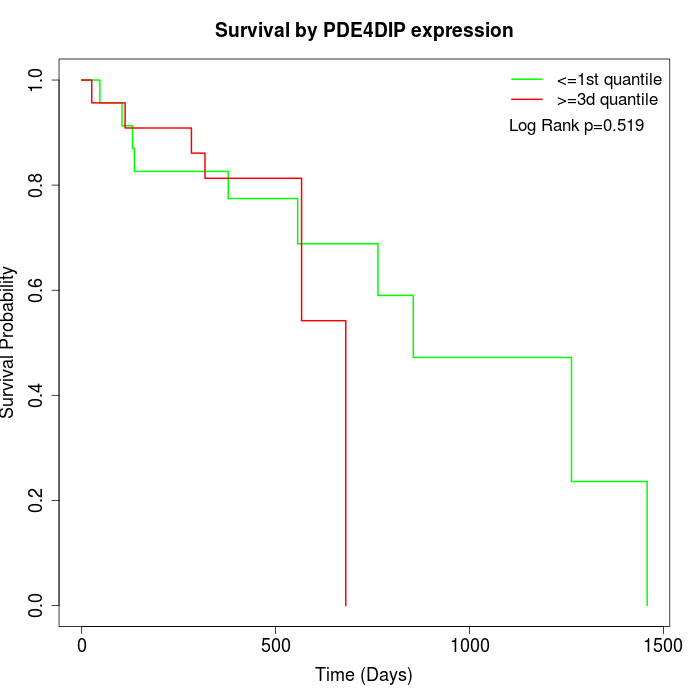
Survival by PDE4DIP expression:
Note: Click image to view full size file.
Copy number change of PDE4DIP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE4DIP | 9659 | 11 | 2 | 17 | |
| GSE20123 | PDE4DIP | 9659 | 10 | 3 | 17 | |
| GSE43470 | PDE4DIP | 9659 | 5 | 2 | 36 | |
| GSE46452 | PDE4DIP | 9659 | 2 | 1 | 56 | |
| GSE47630 | PDE4DIP | 9659 | 12 | 2 | 26 | |
| GSE54993 | PDE4DIP | 9659 | 0 | 2 | 68 | |
| GSE54994 | PDE4DIP | 9659 | 15 | 0 | 38 | |
| GSE60625 | PDE4DIP | 9659 | 0 | 0 | 11 | |
| GSE74703 | PDE4DIP | 9659 | 5 | 2 | 29 | |
| GSE74704 | PDE4DIP | 9659 | 5 | 1 | 14 | |
| TCGA | PDE4DIP | 9659 | 33 | 15 | 48 |
Total number of gains: 98; Total number of losses: 30; Total Number of normals: 360.
Somatic mutations of PDE4DIP:
Generating mutation plots.
Highly correlated genes for PDE4DIP:
Showing top 20/680 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE4DIP | MOAP1 | 0.796416 | 3 | 0 | 3 |
| PDE4DIP | ANO5 | 0.786563 | 3 | 0 | 3 |
| PDE4DIP | CCDC69 | 0.753397 | 5 | 0 | 4 |
| PDE4DIP | AFF3 | 0.749766 | 3 | 0 | 3 |
| PDE4DIP | NEGR1 | 0.74518 | 4 | 0 | 4 |
| PDE4DIP | ANGPTL1 | 0.740289 | 3 | 0 | 3 |
| PDE4DIP | GPR37 | 0.737736 | 3 | 0 | 3 |
| PDE4DIP | HS6ST3 | 0.735533 | 3 | 0 | 3 |
| PDE4DIP | RNF150 | 0.734807 | 4 | 0 | 4 |
| PDE4DIP | FIBIN | 0.723308 | 4 | 0 | 3 |
| PDE4DIP | PSIP1 | 0.717091 | 3 | 0 | 3 |
| PDE4DIP | S1PR3 | 0.71499 | 5 | 0 | 5 |
| PDE4DIP | ASB5 | 0.711678 | 3 | 0 | 3 |
| PDE4DIP | GMPR | 0.70977 | 4 | 0 | 4 |
| PDE4DIP | CBY1 | 0.709148 | 3 | 0 | 3 |
| PDE4DIP | CH25H | 0.70572 | 3 | 0 | 3 |
| PDE4DIP | BNC2 | 0.704324 | 5 | 0 | 5 |
| PDE4DIP | FAM229B | 0.703504 | 3 | 0 | 3 |
| PDE4DIP | KCNB1 | 0.703241 | 3 | 0 | 3 |
| PDE4DIP | FNIP2 | 0.702309 | 4 | 0 | 4 |
For details and further investigation, click here